Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
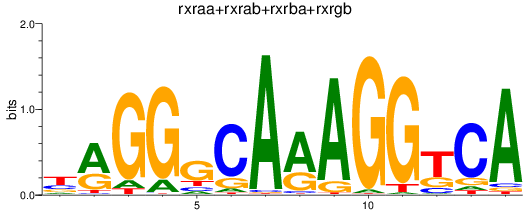
Results for rxraa+rxrab+rxrba+rxrgb
Z-value: 1.14
Transcription factors associated with rxraa+rxrab+rxrba+rxrgb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rxrgb
|
ENSDARG00000004697 | retinoid X receptor, gamma b |
|
rxrab
|
ENSDARG00000035127 | retinoid x receptor, alpha b |
|
rxraa
|
ENSDARG00000057737 | retinoid X receptor, alpha a |
|
rxrba
|
ENSDARG00000078954 | retinoid x receptor, beta a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rxrgb | dr11_v1_chr20_+_33875256_33875256 | 0.48 | 1.0e-06 | Click! |
| rxrba | dr11_v1_chr19_-_7272921_7273057 | -0.38 | 1.5e-04 | Click! |
| rxrab | dr11_v1_chr5_-_64511428_64511428 | -0.24 | 1.9e-02 | Click! |
| rxraa | dr11_v1_chr21_+_17768174_17768174 | 0.18 | 8.2e-02 | Click! |
Activity profile of rxraa+rxrab+rxrba+rxrgb motif
Sorted Z-values of rxraa+rxrab+rxrba+rxrgb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_19006631 | 18.77 |
ENSDART00000080269
ENSDART00000191682 |
pgam2
|
phosphoglycerate mutase 2 (muscle) |
| chr7_+_34297271 | 18.53 |
ENSDART00000180342
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr13_+_50375800 | 17.18 |
ENSDART00000099537
|
cox5b2
|
cytochrome c oxidase subunit Vb 2 |
| chr7_+_34296789 | 14.52 |
ENSDART00000052471
ENSDART00000173798 ENSDART00000173778 |
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr6_+_60055168 | 13.14 |
ENSDART00000008752
|
pck1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr2_-_39017838 | 12.26 |
ENSDART00000048838
|
rbp2b
|
retinol binding protein 2b, cellular |
| chr22_+_38229321 | 11.55 |
ENSDART00000132670
ENSDART00000104504 |
si:ch211-284e20.8
|
si:ch211-284e20.8 |
| chr22_-_7050 | 11.06 |
ENSDART00000127829
|
atad3
|
ATPase family, AAA domain containing 3 |
| chr22_-_23612854 | 10.94 |
ENSDART00000165885
|
cfhl5
|
complement factor H like 5 |
| chr12_-_6033824 | 10.91 |
ENSDART00000131301
ENSDART00000139419 ENSDART00000032050 |
g6pca.1
|
glucose-6-phosphatase a, catalytic subunit, tandem duplicate 1 |
| chr20_+_15015557 | 10.73 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr11_+_11201096 | 10.63 |
ENSDART00000171916
ENSDART00000171521 ENSDART00000087105 ENSDART00000159603 |
myom2a
|
myomesin 2a |
| chr18_-_17415580 | 10.44 |
ENSDART00000150077
ENSDART00000100190 |
ces2
|
carboxylesterase 2 (intestine, liver) |
| chr3_-_18805225 | 9.93 |
ENSDART00000133471
ENSDART00000131758 |
msrb1a
|
methionine sulfoxide reductase B1a |
| chr21_+_20771082 | 9.60 |
ENSDART00000079732
|
oxct1b
|
3-oxoacid CoA transferase 1b |
| chr5_-_36837846 | 9.57 |
ENSDART00000032481
|
ckma
|
creatine kinase, muscle a |
| chr11_+_18037729 | 9.17 |
ENSDART00000111624
|
zgc:175135
|
zgc:175135 |
| chr11_-_22303678 | 8.92 |
ENSDART00000159527
ENSDART00000159681 |
tfeb
|
transcription factor EB |
| chr2_+_42191592 | 8.89 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr7_-_52096498 | 8.85 |
ENSDART00000098688
ENSDART00000098690 |
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr17_+_450956 | 8.79 |
ENSDART00000183022
ENSDART00000171386 |
zgc:194887
|
zgc:194887 |
| chr23_-_12788113 | 8.78 |
ENSDART00000103231
ENSDART00000146640 ENSDART00000180549 |
si:dkey-96f10.1
|
si:dkey-96f10.1 |
| chr18_+_1615 | 8.71 |
ENSDART00000082450
|
homer2
|
homer scaffolding protein 2 |
| chr9_-_22834860 | 8.71 |
ENSDART00000146486
|
neb
|
nebulin |
| chr11_+_18157260 | 8.46 |
ENSDART00000144659
|
zgc:173545
|
zgc:173545 |
| chr8_-_37263524 | 8.44 |
ENSDART00000061327
|
rh50
|
Rh50-like protein |
| chr5_-_69940868 | 8.35 |
ENSDART00000185924
ENSDART00000097357 |
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr20_+_47953047 | 8.13 |
ENSDART00000079734
|
hadhaa
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha a |
| chr11_+_18053333 | 7.99 |
ENSDART00000075750
|
zgc:175135
|
zgc:175135 |
| chr19_-_41213718 | 7.98 |
ENSDART00000077121
|
pdk4
|
pyruvate dehydrogenase kinase, isozyme 4 |
| chr8_-_65189 | 7.62 |
ENSDART00000168412
|
hsd17b4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr11_+_18130300 | 7.61 |
ENSDART00000169146
|
zgc:175135
|
zgc:175135 |
| chr2_+_22659787 | 7.40 |
ENSDART00000043956
|
zgc:161973
|
zgc:161973 |
| chr20_+_54304800 | 7.40 |
ENSDART00000121661
|
zp2.6
|
zona pellucida glycoprotein 2, tandem duplicate 6 |
| chr19_+_40861853 | 7.37 |
ENSDART00000126470
|
zgc:85777
|
zgc:85777 |
| chr20_-_25522911 | 7.35 |
ENSDART00000063058
|
cyp2n13
|
cytochrome P450, family 2, subfamily N, polypeptide 13 |
| chr15_-_6946286 | 7.27 |
ENSDART00000019330
|
ech1
|
enoyl CoA hydratase 1, peroxisomal |
| chr14_-_48103207 | 7.25 |
ENSDART00000056712
|
etfdh
|
electron-transferring-flavoprotein dehydrogenase |
| chr12_-_5120175 | 7.15 |
ENSDART00000160729
|
rbp4
|
retinol binding protein 4, plasma |
| chr20_+_54312970 | 7.09 |
ENSDART00000024598
ENSDART00000193172 |
zp2.5
|
zona pellucida glycoprotein 2, tandem duplicate 5 |
| chr12_-_5120339 | 6.92 |
ENSDART00000168759
|
rbp4
|
retinol binding protein 4, plasma |
| chr3_+_12732382 | 6.84 |
ENSDART00000158403
|
cyp2k19
|
cytochrome P450, family 2, subfamily k, polypeptide 19 |
| chr3_+_39566999 | 6.83 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr20_+_54309148 | 6.79 |
ENSDART00000099360
|
zp2.1
|
zona pellucida glycoprotein 2, tandem duplicate 1 |
| chr21_+_25777425 | 6.70 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr7_+_69019851 | 6.68 |
ENSDART00000162891
|
CABZ01057488.1
|
|
| chr6_-_59563597 | 6.56 |
ENSDART00000166311
|
INHBE
|
inhibin beta E subunit |
| chr20_+_54295213 | 6.51 |
ENSDART00000074085
|
zp2.3
|
zona pellucida glycoprotein 2, tandem duplicate 3 |
| chr20_+_54299419 | 6.44 |
ENSDART00000056089
ENSDART00000193107 |
si:zfos-1505d6.3
|
si:zfos-1505d6.3 |
| chr11_+_77526 | 6.36 |
ENSDART00000193521
|
CABZ01072242.1
|
|
| chr5_-_1962500 | 6.17 |
ENSDART00000150163
|
rplp0
|
ribosomal protein, large, P0 |
| chr7_-_52096287 | 6.11 |
ENSDART00000174068
|
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr13_-_34683370 | 6.04 |
ENSDART00000113661
|
kif16bb
|
kinesin family member 16Bb |
| chr9_+_33216945 | 6.00 |
ENSDART00000134029
|
si:ch211-125e6.12
|
si:ch211-125e6.12 |
| chr22_-_20126230 | 5.99 |
ENSDART00000138688
|
creb3l3a
|
cAMP responsive element binding protein 3-like 3a |
| chr9_-_43082945 | 5.95 |
ENSDART00000142257
|
ccdc141
|
coiled-coil domain containing 141 |
| chr7_-_51953613 | 5.93 |
ENSDART00000142042
|
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr7_-_24046999 | 5.90 |
ENSDART00000144616
ENSDART00000124653 ENSDART00000127813 |
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr8_-_2529878 | 5.88 |
ENSDART00000056767
|
acaa2
|
acetyl-CoA acyltransferase 2 |
| chr22_-_38274188 | 5.84 |
ENSDART00000139420
ENSDART00000015117 |
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr11_+_18175893 | 5.81 |
ENSDART00000177625
|
zgc:173545
|
zgc:173545 |
| chr13_+_829585 | 5.81 |
ENSDART00000029051
|
gsta.2
|
glutathione S-transferase, alpha tandem duplicate 2 |
| chr7_-_17337233 | 5.80 |
ENSDART00000050236
ENSDART00000102141 |
nitr8
|
novel immune-type receptor 8 |
| chr24_-_11446156 | 5.78 |
ENSDART00000143921
ENSDART00000066778 |
acad11
|
acyl-CoA dehydrogenase family, member 11 |
| chr19_+_46113828 | 5.73 |
ENSDART00000159331
ENSDART00000161826 |
rbm24a
|
RNA binding motif protein 24a |
| chr22_-_5171362 | 5.68 |
ENSDART00000124889
|
tnfaip8l1
|
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr2_-_26476030 | 5.61 |
ENSDART00000145262
ENSDART00000132125 |
acadm
|
acyl-CoA dehydrogenase medium chain |
| chr2_-_57076687 | 5.55 |
ENSDART00000161523
|
slc25a42
|
solute carrier family 25, member 42 |
| chr3_+_1749793 | 5.53 |
ENSDART00000149308
|
si:dkeyp-52c3.7
|
si:dkeyp-52c3.7 |
| chr3_+_12744083 | 5.36 |
ENSDART00000158554
ENSDART00000169545 |
cyp2k21
|
cytochrome P450, family 2, subfamily k, polypeptide 21 |
| chr19_-_27564458 | 5.35 |
ENSDART00000123155
|
si:dkeyp-46h3.6
|
si:dkeyp-46h3.6 |
| chr9_+_35077546 | 5.16 |
ENSDART00000142688
|
si:dkey-192g7.3
|
si:dkey-192g7.3 |
| chr20_+_572037 | 5.16 |
ENSDART00000028062
ENSDART00000152736 ENSDART00000031759 ENSDART00000162198 |
smyd2b
|
SET and MYND domain containing 2b |
| chr20_-_47953524 | 5.13 |
ENSDART00000167986
|
HADHB
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit beta |
| chr1_-_9641845 | 5.11 |
ENSDART00000121490
ENSDART00000159411 |
ugt5b2
ugt5b3
|
UDP glucuronosyltransferase 5 family, polypeptide B2 UDP glucuronosyltransferase 5 family, polypeptide B3 |
| chr3_+_27770110 | 4.99 |
ENSDART00000017962
|
eci1
|
enoyl-CoA delta isomerase 1 |
| chr18_+_44769027 | 4.98 |
ENSDART00000145190
|
ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr7_-_24520866 | 4.83 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr24_+_2961098 | 4.80 |
ENSDART00000163760
ENSDART00000170835 |
eci2
|
enoyl-CoA delta isomerase 2 |
| chr16_-_23800484 | 4.80 |
ENSDART00000139964
|
rps27.2
|
ribosomal protein S27, isoform 2 |
| chr17_-_32370047 | 4.71 |
ENSDART00000145487
|
klf11b
|
Kruppel-like factor 11b |
| chr7_+_31871830 | 4.64 |
ENSDART00000139899
|
mybpc3
|
myosin binding protein C, cardiac |
| chr19_+_7043634 | 4.56 |
ENSDART00000133954
|
mhc1uka
|
major histocompatibility complex class I UKA |
| chr7_+_71664624 | 4.54 |
ENSDART00000170273
|
emilin2b
|
elastin microfibril interfacer 2b |
| chr15_-_16884912 | 4.49 |
ENSDART00000062135
|
zgc:103681
|
zgc:103681 |
| chr16_-_19568388 | 4.48 |
ENSDART00000141616
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr1_-_58868306 | 4.42 |
ENSDART00000166615
|
dnm2b
|
dynamin 2b |
| chr3_+_16922226 | 4.42 |
ENSDART00000017646
|
atp6v0a1a
|
ATPase H+ transporting V0 subunit a1a |
| chr12_-_47774807 | 4.41 |
ENSDART00000193831
|
LO017725.1
|
|
| chr19_+_32166702 | 4.36 |
ENSDART00000021798
|
fabp11a
|
fatty acid binding protein 11a |
| chr17_+_51906053 | 4.27 |
ENSDART00000159072
ENSDART00000056869 |
flvcr2a
|
feline leukemia virus subgroup C cellular receptor family, member 2a |
| chr8_+_11687254 | 4.26 |
ENSDART00000042040
|
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr20_-_48604621 | 4.23 |
ENSDART00000161769
ENSDART00000157871 |
mgst3a
|
microsomal glutathione S-transferase 3a |
| chr5_+_43870389 | 4.23 |
ENSDART00000141002
|
zgc:112966
|
zgc:112966 |
| chr5_+_6670945 | 4.22 |
ENSDART00000185686
|
pxna
|
paxillin a |
| chr10_-_690072 | 4.22 |
ENSDART00000164871
ENSDART00000142833 |
glis3
|
GLIS family zinc finger 3 |
| chr11_-_29996344 | 4.21 |
ENSDART00000003712
ENSDART00000126110 |
ace2
|
angiotensin I converting enzyme 2 |
| chr1_-_9644630 | 4.21 |
ENSDART00000123725
ENSDART00000161164 |
ugt5b3
|
UDP glucuronosyltransferase 5 family, polypeptide B3 |
| chr15_+_404891 | 4.17 |
ENSDART00000155682
|
nipsnap2
|
nipsnap homolog 2 |
| chr23_+_42434348 | 4.02 |
ENSDART00000161027
|
cyp2aa1
|
cytochrome P450, family 2, subfamily AA, polypeptide 1 |
| chr21_-_1640547 | 3.99 |
ENSDART00000151041
|
zgc:152948
|
zgc:152948 |
| chr3_-_34561624 | 3.99 |
ENSDART00000129313
|
sept9a
|
septin 9a |
| chr24_-_7587401 | 3.99 |
ENSDART00000093163
|
galnt11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr25_+_36347126 | 3.98 |
ENSDART00000152449
|
si:ch211-113a14.22
|
si:ch211-113a14.22 |
| chr20_-_48604199 | 3.97 |
ENSDART00000161762
ENSDART00000170894 |
mgst3a
|
microsomal glutathione S-transferase 3a |
| chr16_-_24612871 | 3.94 |
ENSDART00000155614
ENSDART00000154787 ENSDART00000155983 ENSDART00000156519 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr23_+_19790962 | 3.93 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr18_+_39487486 | 3.91 |
ENSDART00000126978
|
acadl
|
acyl-CoA dehydrogenase long chain |
| chr20_+_35445462 | 3.90 |
ENSDART00000124497
|
tdrd6
|
tudor domain containing 6 |
| chr1_-_49505449 | 3.90 |
ENSDART00000187294
ENSDART00000132171 |
si:dkeyp-80c12.5
|
si:dkeyp-80c12.5 |
| chr23_-_45705525 | 3.88 |
ENSDART00000148959
|
ednrab
|
endothelin receptor type Ab |
| chr21_-_5879897 | 3.87 |
ENSDART00000184034
|
rpl35
|
ribosomal protein L35 |
| chr23_-_10696626 | 3.86 |
ENSDART00000177571
|
foxp1a
|
forkhead box P1a |
| chr24_+_81527 | 3.86 |
ENSDART00000192139
|
reck
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr6_-_7082108 | 3.83 |
ENSDART00000081760
|
ihhb
|
Indian hedgehog homolog b |
| chr14_+_34495216 | 3.78 |
ENSDART00000147756
|
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr7_+_31838320 | 3.77 |
ENSDART00000144679
ENSDART00000174217 ENSDART00000122506 |
mybpc3
|
myosin binding protein C, cardiac |
| chr6_+_153146 | 3.73 |
ENSDART00000097468
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr21_+_5080789 | 3.73 |
ENSDART00000024199
|
atp5fa1
|
ATP synthase F1 subunit alpha |
| chr23_-_46040618 | 3.68 |
ENSDART00000161415
|
CABZ01080918.1
|
|
| chr3_+_12718100 | 3.68 |
ENSDART00000162343
ENSDART00000192425 |
cyp2k20
|
cytochrome P450, family 2, subfamily k, polypeptide 20 |
| chr17_+_53297822 | 3.66 |
ENSDART00000168297
|
ddx24
|
DEAD (Asp-Glu-Ala-Asp) box helicase 24 |
| chr22_-_26834043 | 3.65 |
ENSDART00000087202
|
si:dkey-44g23.5
|
si:dkey-44g23.5 |
| chr6_+_60036767 | 3.64 |
ENSDART00000155009
|
tgm8
|
transglutaminase 8 |
| chr18_-_15771551 | 3.64 |
ENSDART00000130931
ENSDART00000154079 |
si:ch211-219a15.3
|
si:ch211-219a15.3 |
| chr5_-_36549024 | 3.62 |
ENSDART00000097671
|
zgc:158432
|
zgc:158432 |
| chr23_-_44903048 | 3.61 |
ENSDART00000149103
|
fhdc5
|
FH2 domain containing 5 |
| chr11_+_24313931 | 3.55 |
ENSDART00000017599
ENSDART00000166045 |
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr9_+_8380728 | 3.54 |
ENSDART00000133501
|
si:ch1073-75o15.4
|
si:ch1073-75o15.4 |
| chr11_+_29965822 | 3.48 |
ENSDART00000127049
|
il1fma
|
interleukin-1 family member A |
| chr13_-_25199260 | 3.46 |
ENSDART00000057605
|
adka
|
adenosine kinase a |
| chr3_-_36419641 | 3.43 |
ENSDART00000173545
|
cog1
|
component of oligomeric golgi complex 1 |
| chr21_+_29179887 | 3.43 |
ENSDART00000161941
|
si:ch211-57b15.1
|
si:ch211-57b15.1 |
| chr17_+_30546579 | 3.37 |
ENSDART00000154385
|
nhsl1a
|
NHS-like 1a |
| chr8_+_48491387 | 3.33 |
ENSDART00000086053
|
PRDM16
|
si:ch211-263k4.2 |
| chr7_-_51953807 | 3.32 |
ENSDART00000174102
ENSDART00000145645 ENSDART00000052054 |
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr3_+_25999477 | 3.31 |
ENSDART00000024316
|
mcm5
|
minichromosome maintenance complex component 5 |
| chr2_-_37744951 | 3.30 |
ENSDART00000144807
|
myo9b
|
myosin IXb |
| chr3_+_4346854 | 3.29 |
ENSDART00000004273
|
si:dkey-73p2.3
|
si:dkey-73p2.3 |
| chr13_-_40726865 | 3.29 |
ENSDART00000099847
|
st3gal7
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 7 |
| chr24_-_26820698 | 3.26 |
ENSDART00000147788
|
fndc3bb
|
fibronectin type III domain containing 3Bb |
| chr15_+_5088210 | 3.25 |
ENSDART00000183423
|
mxf
|
myxovirus (influenza virus) resistance F |
| chr15_-_28082310 | 3.24 |
ENSDART00000152620
|
dhrs13a.3
|
dehydrogenase/reductase (SDR family) member 13a, duplicate 3 |
| chr21_+_15704556 | 3.22 |
ENSDART00000024858
ENSDART00000146909 |
chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr19_-_35439237 | 3.18 |
ENSDART00000145883
|
anln
|
anillin, actin binding protein |
| chr23_+_44634187 | 3.18 |
ENSDART00000143688
|
si:ch73-265d7.2
|
si:ch73-265d7.2 |
| chr6_-_48082525 | 3.10 |
ENSDART00000192049
|
slc2a1b
|
solute carrier family 2 (facilitated glucose transporter), member 1b |
| chr21_-_30166097 | 3.10 |
ENSDART00000130676
|
hbegfb
|
heparin-binding EGF-like growth factor b |
| chr4_-_50926767 | 3.10 |
ENSDART00000183430
|
si:ch211-208f21.3
|
si:ch211-208f21.3 |
| chr3_+_58167288 | 3.09 |
ENSDART00000155874
ENSDART00000010395 |
uqcrc2a
|
ubiquinol-cytochrome c reductase core protein 2a |
| chr18_-_34549721 | 3.09 |
ENSDART00000137101
ENSDART00000021880 |
ssr3
|
signal sequence receptor, gamma |
| chr24_+_26742226 | 3.08 |
ENSDART00000079721
|
ghsrb
|
growth hormone secretagogue receptor b |
| chr8_+_32742650 | 3.06 |
ENSDART00000138117
|
hmcn2
|
hemicentin 2 |
| chr6_+_1787160 | 3.05 |
ENSDART00000113505
|
myl9b
|
myosin, light chain 9b, regulatory |
| chr12_-_36268723 | 3.05 |
ENSDART00000113740
|
kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr16_-_1503023 | 3.04 |
ENSDART00000036348
|
sim1a
|
single-minded family bHLH transcription factor 1a |
| chr15_+_28268135 | 3.03 |
ENSDART00000152536
ENSDART00000188550 |
myo1cb
|
myosin Ic, paralog b |
| chr5_-_30984271 | 3.02 |
ENSDART00000051392
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr2_+_57801960 | 3.01 |
ENSDART00000147966
|
si:dkeyp-68b7.10
|
si:dkeyp-68b7.10 |
| chr13_+_844150 | 2.99 |
ENSDART00000058260
|
gsta.1
|
glutathione S-transferase, alpha tandem duplicate 1 |
| chr2_-_24289641 | 2.99 |
ENSDART00000128784
ENSDART00000123565 ENSDART00000141922 ENSDART00000184550 ENSDART00000191469 |
myh7l
|
myosin heavy chain 7-like |
| chr22_+_18469004 | 2.98 |
ENSDART00000061430
|
cilp2
|
cartilage intermediate layer protein 2 |
| chr8_+_30664077 | 2.98 |
ENSDART00000138750
|
adora2aa
|
adenosine A2a receptor a |
| chr17_+_6538733 | 2.97 |
ENSDART00000193712
|
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr16_+_28602423 | 2.97 |
ENSDART00000182350
|
crot
|
carnitine O-octanoyltransferase |
| chr1_-_56537347 | 2.97 |
ENSDART00000126054
|
CABZ01059406.1
|
|
| chr11_-_45135643 | 2.91 |
ENSDART00000170863
|
cant1b
|
calcium activated nucleotidase 1b |
| chr23_-_16737161 | 2.85 |
ENSDART00000132573
|
si:ch211-224l10.4
|
si:ch211-224l10.4 |
| chr7_+_24520518 | 2.84 |
ENSDART00000173604
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr5_-_30984010 | 2.81 |
ENSDART00000182367
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr18_+_13315739 | 2.81 |
ENSDART00000143404
|
si:ch211-260p9.3
|
si:ch211-260p9.3 |
| chr3_-_55531450 | 2.81 |
ENSDART00000155376
|
tex2
|
testis expressed 2 |
| chr22_-_17595310 | 2.80 |
ENSDART00000099056
|
gpx4a
|
glutathione peroxidase 4a |
| chr8_+_31435452 | 2.73 |
ENSDART00000145282
|
selenop
|
selenoprotein P |
| chr10_+_11265387 | 2.71 |
ENSDART00000038888
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr3_+_1179601 | 2.70 |
ENSDART00000173378
|
triobpb
|
TRIO and F-actin binding protein b |
| chr15_+_15403560 | 2.69 |
ENSDART00000049831
|
dhrs11b
|
dehydrogenase/reductase (SDR family) member 11b |
| chr23_-_1587955 | 2.67 |
ENSDART00000136037
|
fndc7b
|
fibronectin type III domain containing 7b |
| chr1_+_56955349 | 2.67 |
ENSDART00000152758
|
si:ch211-1f22.10
|
si:ch211-1f22.10 |
| chr13_-_28688104 | 2.64 |
ENSDART00000133827
|
pcgf6
|
polycomb group ring finger 6 |
| chr1_+_57145072 | 2.61 |
ENSDART00000152776
|
si:ch73-94k4.4
|
si:ch73-94k4.4 |
| chr5_+_42141917 | 2.60 |
ENSDART00000172201
ENSDART00000140743 |
trpv1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chr4_-_14531687 | 2.60 |
ENSDART00000182093
ENSDART00000159447 |
plxnb2a
|
plexin b2a |
| chr14_-_28052474 | 2.60 |
ENSDART00000172948
ENSDART00000135337 |
TSC22D3 (1 of many)
zgc:64189
|
si:ch211-220e11.3 zgc:64189 |
| chr22_-_14262115 | 2.55 |
ENSDART00000168264
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr1_-_46859398 | 2.54 |
ENSDART00000135795
|
agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr22_+_38778649 | 2.53 |
ENSDART00000075873
|
alpi.2
|
alkaline phosphatase, intestinal, tandem duplicate 2 |
| chr2_-_31686353 | 2.53 |
ENSDART00000126177
ENSDART00000056679 |
e2f5
|
E2F transcription factor 5 |
| chr12_+_1286642 | 2.50 |
ENSDART00000157467
|
pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr4_-_31064105 | 2.48 |
ENSDART00000157670
|
si:dkey-11n14.1
|
si:dkey-11n14.1 |
| chr1_+_57176256 | 2.48 |
ENSDART00000152741
|
si:dkey-27j5.6
|
si:dkey-27j5.6 |
| chr3_+_3810919 | 2.47 |
ENSDART00000056035
|
FQ311927.1
|
|
| chr5_+_38631687 | 2.47 |
ENSDART00000147280
|
si:ch211-271e10.6
|
si:ch211-271e10.6 |
| chr17_-_12196865 | 2.47 |
ENSDART00000154694
|
kif28
|
kinesin family member 28 |
| chr21_+_14511685 | 2.46 |
ENSDART00000102099
|
CR392036.1
|
|
| chr8_+_999421 | 2.46 |
ENSDART00000149528
|
fabp1b.1
|
fatty acid binding protein 1b, tandem duplicate 1 |
| chr8_-_4097722 | 2.45 |
ENSDART00000135006
|
cux2b
|
cut-like homeobox 2b |
| chr24_-_25186895 | 2.42 |
ENSDART00000081029
|
plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr1_+_58312187 | 2.39 |
ENSDART00000142285
|
si:dkey-222h21.10
|
si:dkey-222h21.10 |
| chr14_+_30340251 | 2.38 |
ENSDART00000148448
|
mtus1a
|
microtubule associated tumor suppressor 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of rxraa+rxrab+rxrba+rxrgb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 34.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 3.3 | 13.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 2.4 | 7.3 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 2.0 | 14.1 | GO:0034633 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 1.9 | 9.6 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 1.7 | 8.4 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 1.7 | 6.6 | GO:0006549 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 1.5 | 8.7 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 1.3 | 6.7 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 1.3 | 17.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 1.3 | 3.9 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 1.3 | 3.8 | GO:0021512 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 1.2 | 3.5 | GO:0046689 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 1.1 | 8.7 | GO:0021627 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 1.1 | 4.3 | GO:0097037 | heme export(GO:0097037) |
| 1.1 | 8.4 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 1.0 | 3.0 | GO:0072020 | proximal straight tubule development(GO:0072020) |
| 1.0 | 38.1 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.9 | 1.8 | GO:0055057 | neuroblast division(GO:0055057) |
| 0.9 | 44.3 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.8 | 3.2 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.7 | 3.0 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.7 | 3.0 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) vitamin transmembrane transport(GO:0035461) coenzyme transport(GO:0051182) |
| 0.7 | 3.7 | GO:0035678 | neuromast hair cell morphogenesis(GO:0035678) |
| 0.7 | 18.8 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.7 | 3.5 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.7 | 11.1 | GO:0001952 | regulation of cell-matrix adhesion(GO:0001952) |
| 0.6 | 3.9 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.6 | 5.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.6 | 3.8 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.6 | 3.5 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.6 | 1.7 | GO:1903961 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.5 | 8.8 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.5 | 2.7 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.4 | 1.8 | GO:0003161 | cardiac conduction system development(GO:0003161) negative regulation of heart rate(GO:0010459) |
| 0.4 | 8.9 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.4 | 1.3 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.4 | 3.1 | GO:0043589 | skin morphogenesis(GO:0043589) mesenchymal cell migration(GO:0090497) |
| 0.4 | 7.7 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.4 | 1.2 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.4 | 5.7 | GO:0061055 | myotome development(GO:0061055) |
| 0.4 | 3.7 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.4 | 3.9 | GO:0030719 | P granule organization(GO:0030719) |
| 0.3 | 3.1 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.3 | 4.4 | GO:0001845 | phagolysosome assembly(GO:0001845) |
| 0.3 | 2.7 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.3 | 4.3 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.3 | 1.0 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.3 | 3.0 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.3 | 6.8 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.3 | 1.3 | GO:0046551 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.3 | 2.5 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.3 | 3.3 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.3 | 5.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.3 | 1.6 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.3 | 1.9 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.3 | 1.4 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.3 | 6.2 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.3 | 2.3 | GO:0006833 | water transport(GO:0006833) |
| 0.3 | 2.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.2 | 14.3 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.2 | 0.7 | GO:0019408 | dolichol biosynthetic process(GO:0019408) |
| 0.2 | 0.7 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.2 | 1.8 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.2 | 1.8 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.2 | 24.5 | GO:0048545 | response to steroid hormone(GO:0048545) |
| 0.2 | 6.5 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.2 | 2.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.2 | 8.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.2 | 4.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.2 | 1.4 | GO:0048662 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.2 | 0.6 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.2 | 5.5 | GO:0048599 | oocyte development(GO:0048599) |
| 0.2 | 2.6 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.2 | 3.1 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.2 | 1.9 | GO:0021681 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 | 3.2 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.2 | 1.8 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.2 | 0.8 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.2 | 2.0 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.2 | 1.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 3.7 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.4 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 5.7 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 11.6 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 1.3 | GO:0060337 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 2.2 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.1 | 2.6 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 6.0 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.1 | 4.9 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 2.9 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 1.7 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.2 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 1.7 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 2.2 | GO:0051058 | negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.1 | 10.3 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 0.5 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.1 | 3.1 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.1 | 4.3 | GO:0010675 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.1 | 1.8 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.1 | 1.9 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 7.4 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.1 | 0.5 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.1 | 3.2 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.1 | 5.2 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.1 | 0.6 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 1.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 4.3 | GO:0007006 | mitochondrial membrane organization(GO:0007006) |
| 0.1 | 1.4 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 0.3 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 4.3 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.1 | 1.5 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 4.5 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.1 | 0.3 | GO:1990168 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 2.3 | GO:0043491 | protein kinase B signaling(GO:0043491) |
| 0.1 | 6.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 4.1 | GO:1990266 | neutrophil migration(GO:1990266) |
| 0.1 | 1.0 | GO:0001508 | action potential(GO:0001508) |
| 0.1 | 4.3 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.6 | GO:0035065 | regulation of histone acetylation(GO:0035065) |
| 0.1 | 1.6 | GO:0001878 | response to yeast(GO:0001878) |
| 0.1 | 1.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 10.7 | GO:0030041 | actin filament polymerization(GO:0030041) |
| 0.1 | 4.0 | GO:0008593 | regulation of Notch signaling pathway(GO:0008593) |
| 0.1 | 0.5 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 2.2 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 1.0 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 7.4 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 2.1 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 1.6 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 2.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 10.7 | GO:0007005 | mitochondrion organization(GO:0007005) |
| 0.0 | 0.4 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.5 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.0 | 0.3 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 1.0 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 7.4 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 2.2 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 3.5 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.0 | 1.9 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 1.8 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 5.8 | GO:0009617 | response to bacterium(GO:0009617) |
| 0.0 | 1.5 | GO:0048741 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 7.7 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 1.4 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 2.3 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.9 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 5.7 | GO:0043066 | negative regulation of apoptotic process(GO:0043066) |
| 0.0 | 1.1 | GO:0032880 | regulation of protein localization(GO:0032880) |
| 0.0 | 4.3 | GO:0006955 | immune response(GO:0006955) |
| 0.0 | 0.1 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.5 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 0.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.6 | GO:0005948 | acetolactate synthase complex(GO:0005948) |
| 2.2 | 8.8 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 1.3 | 3.9 | GO:0033391 | chromatoid body(GO:0033391) |
| 1.3 | 3.8 | GO:0097189 | apoptotic body(GO:0097189) |
| 1.0 | 3.9 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.5 | 3.7 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.5 | 10.6 | GO:0031430 | M band(GO:0031430) |
| 0.5 | 3.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.4 | 4.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.4 | 1.6 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.4 | 7.7 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.3 | 5.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.3 | 10.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.3 | 2.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.3 | 0.9 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.3 | 2.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 3.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.2 | 4.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 1.6 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.2 | 0.6 | GO:0030689 | Noc complex(GO:0030689) |
| 0.2 | 3.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 0.7 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.2 | 10.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 1.8 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 6.2 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 23.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 7.3 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 13.4 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 112.6 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.1 | 2.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 4.8 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.1 | 4.0 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 3.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 4.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 0.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 7.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 10.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 8.9 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 3.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 1.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 6.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 1.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 5.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 90.6 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 2.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.0 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 7.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 2.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 3.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.4 | GO:0014069 | postsynaptic density(GO:0014069) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.0 | 33.0 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 3.8 | 18.8 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 2.7 | 8.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 2.5 | 7.6 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 2.5 | 9.9 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 2.4 | 7.3 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 2.2 | 6.6 | GO:0003984 | acetolactate synthase activity(GO:0003984) |
| 2.0 | 14.1 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 2.0 | 5.9 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 1.9 | 9.6 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 1.9 | 5.7 | GO:1990825 | mRNA CDS binding(GO:1990715) sequence-specific mRNA binding(GO:1990825) |
| 1.8 | 5.5 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 1.5 | 9.0 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 1.3 | 3.9 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 1.2 | 5.0 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 1.1 | 3.3 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 1.1 | 13.0 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 1.0 | 6.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 1.0 | 8.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.9 | 3.7 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.9 | 6.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.8 | 14.3 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.8 | 8.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.8 | 49.7 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.7 | 3.0 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.7 | 3.5 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.6 | 3.2 | GO:0016415 | octanoyltransferase activity(GO:0016415) |
| 0.6 | 8.8 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.6 | 17.2 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.6 | 3.9 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.5 | 2.7 | GO:0008430 | selenium binding(GO:0008430) |
| 0.5 | 4.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.5 | 17.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.4 | 2.2 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.4 | 3.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.4 | 1.3 | GO:0043878 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) |
| 0.4 | 3.8 | GO:0005113 | patched binding(GO:0005113) |
| 0.4 | 3.0 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.4 | 3.9 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.3 | 13.1 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.3 | 2.0 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.3 | 8.4 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.3 | 2.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.3 | 3.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.3 | 5.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.3 | 2.5 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.3 | 3.3 | GO:0043142 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.3 | 3.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 4.4 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.3 | 11.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.3 | 4.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.3 | 4.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 2.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.2 | 13.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 1.2 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.2 | 1.3 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.2 | 4.8 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.2 | 1.4 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.2 | 1.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 2.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 6.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 0.7 | GO:0045547 | dehydrodolichyl diphosphate synthase activity(GO:0045547) |
| 0.2 | 2.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.2 | 2.4 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.2 | 1.7 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.2 | 4.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 1.7 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 1.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 1.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 3.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 7.4 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 3.9 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 2.9 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 4.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 1.4 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.1 | 1.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.9 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 3.7 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 1.9 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 3.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 1.0 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.7 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 2.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 1.8 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 0.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 1.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.5 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 4.8 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 5.9 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 1.3 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.1 | 1.8 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 5.8 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 1.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.6 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 1.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 3.7 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.4 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 6.8 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 11.2 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 7.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 8.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 2.9 | GO:0051427 | hormone receptor binding(GO:0051427) |
| 0.0 | 0.3 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.0 | 2.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 4.7 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.7 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 4.0 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 1.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.7 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 1.3 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 5.7 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 1.7 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.9 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 1.4 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 1.0 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 1.9 | GO:0008194 | UDP-glycosyltransferase activity(GO:0008194) |
| 0.0 | 0.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 1.3 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 21.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 2.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 13.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 3.9 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 2.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 2.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 2.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 1.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 10.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 1.4 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.9 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 2.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 13.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 1.4 | 19.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 1.1 | 7.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.9 | 18.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.8 | 6.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.7 | 8.0 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.4 | 3.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.3 | 8.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 1.4 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.3 | 4.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 3.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 1.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 2.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 33.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.2 | 3.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 3.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 1.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 2.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 4.7 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 2.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 4.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 5.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 10.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 2.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.9 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 2.5 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.1 | 2.3 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |