Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
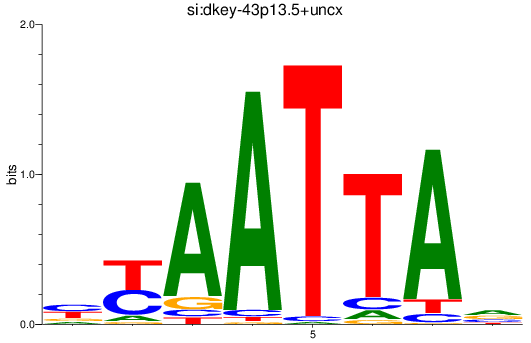
Results for si:dkey-43p13.5+uncx
Z-value: 0.85
Transcription factors associated with si:dkey-43p13.5+uncx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
uncx
|
ENSDARG00000102976 | UNC homeobox |
|
si_dkey-43p13.5
|
ENSDARG00000104199 | si_dkey-43p13.5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| uncx | dr11_v1_chr3_-_43356082_43356082 | -0.71 | 9.6e-16 | Click! |
| si:dkey-43p13.5 | dr11_v1_chr3_+_43086548_43086548 | 0.25 | 1.7e-02 | Click! |
Activity profile of si:dkey-43p13.5+uncx motif
Sorted Z-values of si:dkey-43p13.5+uncx motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_31276842 | 18.42 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr24_+_38301080 | 17.27 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr16_-_17197546 | 11.09 |
ENSDART00000139939
ENSDART00000135146 ENSDART00000063800 ENSDART00000163606 |
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr25_+_31277415 | 9.83 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr21_+_20383837 | 8.36 |
ENSDART00000026430
|
hspb11
|
heat shock protein, alpha-crystallin-related, b11 |
| chr24_-_2450597 | 7.70 |
ENSDART00000188080
ENSDART00000093331 |
rreb1a
|
ras responsive element binding protein 1a |
| chr21_+_28445052 | 7.30 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr14_+_51056605 | 7.18 |
ENSDART00000159639
|
CABZ01078593.1
|
|
| chr25_+_29160102 | 6.71 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr6_+_41191482 | 6.44 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr13_+_22476742 | 6.35 |
ENSDART00000078759
ENSDART00000130101 ENSDART00000137220 ENSDART00000133065 ENSDART00000147348 |
ldb3a
|
LIM domain binding 3a |
| chr21_+_25777425 | 6.26 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr2_+_16781015 | 6.24 |
ENSDART00000155147
ENSDART00000003845 |
tfa
|
transferrin-a |
| chr19_-_5699703 | 6.24 |
ENSDART00000082050
|
zgc:174904
|
zgc:174904 |
| chr3_+_18398876 | 6.18 |
ENSDART00000141100
ENSDART00000138107 |
rps2
|
ribosomal protein S2 |
| chr12_+_3078221 | 6.05 |
ENSDART00000148835
ENSDART00000149427 |
sgca
|
sarcoglycan, alpha |
| chr23_+_39695827 | 6.01 |
ENSDART00000113893
ENSDART00000186679 |
tmco4
|
transmembrane and coiled-coil domains 4 |
| chr10_-_21362320 | 5.82 |
ENSDART00000189789
|
avd
|
avidin |
| chr9_+_8396755 | 5.76 |
ENSDART00000043067
|
zgc:171776
|
zgc:171776 |
| chr16_+_23431189 | 5.73 |
ENSDART00000004679
|
icn
|
ictacalcin |
| chr21_-_5205617 | 5.64 |
ENSDART00000145554
ENSDART00000045284 |
rpl37
|
ribosomal protein L37 |
| chr12_-_35830625 | 5.59 |
ENSDART00000180028
|
CU459056.1
|
|
| chr22_-_31060579 | 5.50 |
ENSDART00000182376
|
cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr25_+_31267268 | 5.39 |
ENSDART00000181239
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr18_+_402048 | 5.34 |
ENSDART00000166345
|
gpib
|
glucose-6-phosphate isomerase b |
| chr10_-_21362071 | 5.27 |
ENSDART00000125167
|
avd
|
avidin |
| chr20_-_22476255 | 5.18 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr9_-_22822084 | 5.15 |
ENSDART00000142020
|
neb
|
nebulin |
| chr24_-_37640705 | 5.09 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr21_+_6556635 | 5.02 |
ENSDART00000139598
|
col5a1
|
procollagen, type V, alpha 1 |
| chr25_+_20089986 | 4.91 |
ENSDART00000143441
ENSDART00000184073 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr20_-_23426339 | 4.83 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr10_+_21867307 | 4.78 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr10_-_8053753 | 4.78 |
ENSDART00000162289
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr7_+_7048245 | 4.77 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr16_+_29509133 | 4.73 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr24_-_40744672 | 4.65 |
ENSDART00000160672
|
CU633479.1
|
|
| chr1_+_135903 | 4.63 |
ENSDART00000124837
|
f10
|
coagulation factor X |
| chr18_+_45781115 | 4.59 |
ENSDART00000151699
ENSDART00000179887 |
rpl35a
|
ribosomal protein L35a |
| chr25_+_3306620 | 4.54 |
ENSDART00000182085
ENSDART00000034704 |
slc25a3b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3b |
| chr5_+_32247310 | 4.52 |
ENSDART00000182649
|
myha
|
myosin, heavy chain a |
| chr20_-_40755614 | 4.45 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr10_+_6884627 | 4.44 |
ENSDART00000125262
ENSDART00000121729 ENSDART00000105384 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr11_+_18183220 | 4.35 |
ENSDART00000113468
|
LO018315.10
|
|
| chr11_-_6452444 | 4.32 |
ENSDART00000137879
ENSDART00000134957 ENSDART00000004483 |
larp6b
|
La ribonucleoprotein domain family, member 6b |
| chr22_-_15593824 | 4.20 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr7_+_20471315 | 4.18 |
ENSDART00000173714
|
si:dkey-19b23.13
|
si:dkey-19b23.13 |
| chr14_+_22113331 | 4.15 |
ENSDART00000109759
|
tmx2a
|
thioredoxin-related transmembrane protein 2a |
| chr23_+_44374041 | 4.07 |
ENSDART00000136056
|
ephb4b
|
eph receptor B4b |
| chr3_+_17537352 | 4.07 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr24_+_12835935 | 4.06 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr10_-_22803740 | 4.03 |
ENSDART00000079469
ENSDART00000187968 ENSDART00000122543 |
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr18_-_19456269 | 4.01 |
ENSDART00000060363
|
rpl4
|
ribosomal protein L4 |
| chr6_-_46861676 | 3.94 |
ENSDART00000188712
ENSDART00000190148 |
igfn1.3
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 3 |
| chr9_-_712308 | 3.93 |
ENSDART00000144625
|
ndufb3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3 |
| chr13_-_20381485 | 3.89 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr19_-_5669122 | 3.88 |
ENSDART00000112211
|
si:ch211-264f5.2
|
si:ch211-264f5.2 |
| chr21_-_1012269 | 3.85 |
ENSDART00000159835
|
CABZ01057159.1
|
|
| chr6_-_54826061 | 3.82 |
ENSDART00000149982
|
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr7_-_51773166 | 3.81 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr1_-_10071422 | 3.78 |
ENSDART00000135522
ENSDART00000033118 |
fga
|
fibrinogen alpha chain |
| chr2_+_24304854 | 3.77 |
ENSDART00000078972
|
fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr7_+_56098590 | 3.73 |
ENSDART00000098453
|
cdh15
|
cadherin 15, type 1, M-cadherin (myotubule) |
| chr10_-_7756865 | 3.69 |
ENSDART00000114373
ENSDART00000125407 ENSDART00000016317 |
loxa
|
lysyl oxidase a |
| chr22_-_10459880 | 3.68 |
ENSDART00000064801
|
ogn
|
osteoglycin |
| chr12_-_26415499 | 3.66 |
ENSDART00000185779
|
synpo2lb
|
synaptopodin 2-like b |
| chr9_-_22821901 | 3.66 |
ENSDART00000101711
|
neb
|
nebulin |
| chr24_-_25166720 | 3.64 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr22_+_19552987 | 3.63 |
ENSDART00000105315
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr6_+_8626427 | 3.60 |
ENSDART00000193660
|
tspeara
|
thrombospondin-type laminin G domain and EAR repeats a |
| chr8_+_45334255 | 3.60 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr21_-_11654422 | 3.60 |
ENSDART00000081614
ENSDART00000132699 |
cast
|
calpastatin |
| chr15_-_23376541 | 3.55 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr10_+_11261576 | 3.55 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr3_-_33427803 | 3.53 |
ENSDART00000075495
|
rpl23
|
ribosomal protein L23 |
| chr24_-_9997948 | 3.50 |
ENSDART00000136274
|
si:ch211-146l10.7
|
si:ch211-146l10.7 |
| chr23_+_42810055 | 3.48 |
ENSDART00000186647
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr5_+_6672870 | 3.47 |
ENSDART00000126598
|
pxna
|
paxillin a |
| chr13_-_12602920 | 3.47 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr24_-_27452488 | 3.46 |
ENSDART00000136433
|
ccl34b.8
|
chemokine (C-C motif) ligand 34b, duplicate 8 |
| chr18_+_39487486 | 3.40 |
ENSDART00000126978
|
acadl
|
acyl-CoA dehydrogenase long chain |
| chr8_+_23738122 | 3.37 |
ENSDART00000062983
|
rpl10a
|
ribosomal protein L10a |
| chr20_-_42702832 | 3.34 |
ENSDART00000134689
ENSDART00000045816 |
plg
|
plasminogen |
| chr12_+_20352400 | 3.33 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr13_+_7442023 | 3.30 |
ENSDART00000080975
|
tnfaip2b
|
tumor necrosis factor, alpha-induced protein 2b |
| chr12_+_10631266 | 3.30 |
ENSDART00000161455
|
csf3a
|
colony stimulating factor 3 (granulocyte) a |
| chr25_-_27621268 | 3.29 |
ENSDART00000146205
ENSDART00000073511 |
hyal6
|
hyaluronoglucosaminidase 6 |
| chr16_-_29387215 | 3.27 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr22_+_15624371 | 3.24 |
ENSDART00000124868
|
lpl
|
lipoprotein lipase |
| chr22_+_1170294 | 3.22 |
ENSDART00000159761
ENSDART00000169809 |
irf6
|
interferon regulatory factor 6 |
| chr25_-_3867990 | 3.16 |
ENSDART00000075663
|
cracr2b
|
calcium release activated channel regulator 2B |
| chr25_+_11456696 | 3.16 |
ENSDART00000171408
|
AGBL1
|
si:ch73-141f14.1 |
| chr17_-_14876758 | 3.14 |
ENSDART00000155857
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr25_+_31405266 | 3.13 |
ENSDART00000103395
|
tnnt3a
|
troponin T type 3a (skeletal, fast) |
| chr16_-_38118003 | 3.11 |
ENSDART00000058667
|
si:dkey-23o4.6
|
si:dkey-23o4.6 |
| chr25_-_3830272 | 3.11 |
ENSDART00000055843
|
cd151
|
CD151 molecule |
| chr22_-_10586191 | 3.10 |
ENSDART00000148418
|
si:dkey-42i9.16
|
si:dkey-42i9.16 |
| chr15_-_21877726 | 3.10 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr2_+_22851832 | 3.09 |
ENSDART00000145944
|
amotl2b
|
angiomotin like 2b |
| chr2_-_51794472 | 3.05 |
ENSDART00000186652
|
BX908782.3
|
|
| chr21_-_22737228 | 3.03 |
ENSDART00000151366
|
fbxo40.2
|
F-box protein 40, tandem duplicate 2 |
| chr10_-_25217347 | 3.03 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr14_+_17376940 | 3.00 |
ENSDART00000054590
ENSDART00000010148 |
spon2b
|
spondin 2b, extracellular matrix protein |
| chr6_-_19271210 | 3.00 |
ENSDART00000163628
ENSDART00000159124 |
zgc:174863
|
zgc:174863 |
| chr13_+_35637048 | 2.99 |
ENSDART00000085037
|
thbs2a
|
thrombospondin 2a |
| chr3_-_29899172 | 2.98 |
ENSDART00000140518
ENSDART00000020311 |
rpl27
|
ribosomal protein L27 |
| chr10_+_8690936 | 2.97 |
ENSDART00000144218
|
si:dkey-27b3.4
|
si:dkey-27b3.4 |
| chr22_-_37686966 | 2.97 |
ENSDART00000192217
|
htr2b
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr1_-_18811517 | 2.95 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr22_-_20695237 | 2.95 |
ENSDART00000112722
|
org
|
oogenesis-related gene |
| chr7_+_20503344 | 2.95 |
ENSDART00000157699
|
si:dkey-19b23.12
|
si:dkey-19b23.12 |
| chr3_-_4663602 | 2.93 |
ENSDART00000083532
|
slc25a38a
|
solute carrier family 25, member 38a |
| chr7_+_19495379 | 2.92 |
ENSDART00000180514
|
si:ch211-212k18.8
|
si:ch211-212k18.8 |
| chr18_+_7204378 | 2.91 |
ENSDART00000142905
|
vwf
|
von Willebrand factor |
| chr1_-_513762 | 2.91 |
ENSDART00000148162
ENSDART00000144606 |
trmt10c
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr6_+_23887314 | 2.89 |
ENSDART00000163188
|
znf648
|
zinc finger protein 648 |
| chr12_+_48803098 | 2.89 |
ENSDART00000074768
|
ppifb
|
peptidylprolyl isomerase Fb |
| chr14_+_35428152 | 2.89 |
ENSDART00000172597
|
sytl4
|
synaptotagmin-like 4 |
| chr18_+_15644559 | 2.87 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr6_+_2097690 | 2.87 |
ENSDART00000193770
|
tgm2b
|
transglutaminase 2b |
| chr13_-_36798204 | 2.87 |
ENSDART00000012357
|
sav1
|
salvador family WW domain containing protein 1 |
| chr12_-_14143344 | 2.86 |
ENSDART00000152742
|
buc2l
|
bucky ball 2-like |
| chr10_-_8046764 | 2.86 |
ENSDART00000099031
|
zgc:136254
|
zgc:136254 |
| chr6_+_11681011 | 2.84 |
ENSDART00000151447
ENSDART00000151618 |
calcrlb
|
calcitonin receptor-like b |
| chr6_-_54815886 | 2.83 |
ENSDART00000180793
ENSDART00000007498 |
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr12_-_18577983 | 2.81 |
ENSDART00000193262
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr11_+_16216909 | 2.81 |
ENSDART00000081035
ENSDART00000147190 |
slc25a26
|
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
| chr22_+_19553390 | 2.79 |
ENSDART00000061739
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr14_+_26247319 | 2.78 |
ENSDART00000192793
|
CCDC69
|
coiled-coil domain containing 69 |
| chr1_-_33645967 | 2.78 |
ENSDART00000192758
|
cldng
|
claudin g |
| chr16_-_15387459 | 2.77 |
ENSDART00000053754
|
has2
|
hyaluronan synthase 2 |
| chr22_-_547748 | 2.76 |
ENSDART00000037455
ENSDART00000140101 |
ccnd3
|
cyclin D3 |
| chr23_-_19686791 | 2.75 |
ENSDART00000161973
|
zgc:193598
|
zgc:193598 |
| chr18_+_33264609 | 2.74 |
ENSDART00000050639
|
v2ra20
|
vomeronasal 2 receptor, a20 |
| chr19_+_46158078 | 2.73 |
ENSDART00000183933
ENSDART00000164055 |
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr5_+_6954162 | 2.73 |
ENSDART00000086666
|
stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr14_+_34492288 | 2.72 |
ENSDART00000144301
|
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr15_-_29598679 | 2.70 |
ENSDART00000155153
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr5_-_7199998 | 2.67 |
ENSDART00000167316
|
CABZ01087502.1
|
|
| chr19_+_43297546 | 2.66 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr4_+_7841627 | 2.65 |
ENSDART00000037997
|
ucmaa
|
upper zone of growth plate and cartilage matrix associated a |
| chr25_+_5972690 | 2.65 |
ENSDART00000067517
|
si:ch211-11i22.4
|
si:ch211-11i22.4 |
| chr6_+_50381665 | 2.64 |
ENSDART00000141128
|
cyc1
|
cytochrome c-1 |
| chr3_-_23643751 | 2.64 |
ENSDART00000078425
ENSDART00000140264 |
eve1
|
even-skipped-like1 |
| chr5_-_1203455 | 2.64 |
ENSDART00000172177
|
surf4
|
surfeit gene 4 |
| chr19_-_25113660 | 2.64 |
ENSDART00000035538
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr7_+_54642005 | 2.62 |
ENSDART00000171864
|
fgf19
|
fibroblast growth factor 19 |
| chr8_-_20230559 | 2.62 |
ENSDART00000193677
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr21_-_19314618 | 2.61 |
ENSDART00000188744
|
gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr18_+_50880096 | 2.61 |
ENSDART00000169782
|
si:ch1073-450f2.1
|
si:ch1073-450f2.1 |
| chr13_+_25486608 | 2.59 |
ENSDART00000057689
|
bag3
|
BCL2 associated athanogene 3 |
| chr19_+_45962016 | 2.58 |
ENSDART00000169710
|
utp23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr4_-_9852318 | 2.58 |
ENSDART00000080702
|
glt8d2
|
glycosyltransferase 8 domain containing 2 |
| chr12_+_22580579 | 2.57 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr20_-_6532462 | 2.56 |
ENSDART00000054653
|
mcm3l
|
MCM3 minichromosome maintenance deficient 3 (S. cerevisiae), like |
| chr5_+_71802014 | 2.55 |
ENSDART00000124939
ENSDART00000097164 |
LHX3
|
LIM homeobox 3 |
| chr22_-_10121880 | 2.55 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr23_-_31913069 | 2.54 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr6_-_3982783 | 2.54 |
ENSDART00000171944
|
slc25a12
|
solute carrier family 25 (aspartate/glutamate carrier), member 12 |
| chr4_+_77943184 | 2.53 |
ENSDART00000159094
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr13_+_28701233 | 2.51 |
ENSDART00000135931
|
si:ch211-67n3.9
|
si:ch211-67n3.9 |
| chr23_-_24394719 | 2.51 |
ENSDART00000044918
|
epha2b
|
eph receptor A2 b |
| chr23_-_32162810 | 2.49 |
ENSDART00000155905
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr16_-_42056137 | 2.48 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr21_-_293146 | 2.48 |
ENSDART00000157781
|
plgrkt
|
plasminogen receptor, C-terminal lysine transmembrane protein |
| chr16_+_33902006 | 2.48 |
ENSDART00000161807
ENSDART00000159474 |
gnl2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr16_+_23913943 | 2.47 |
ENSDART00000175404
ENSDART00000129525 |
apoa4b.1
|
apolipoprotein A-IV b, tandem duplicate 1 |
| chr6_-_39270851 | 2.44 |
ENSDART00000148839
|
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr19_+_15441022 | 2.43 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr20_-_9095105 | 2.43 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr2_+_3201345 | 2.43 |
ENSDART00000130349
|
wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr11_+_36665359 | 2.42 |
ENSDART00000166144
|
si:ch211-11c3.9
|
si:ch211-11c3.9 |
| chr7_-_38658411 | 2.42 |
ENSDART00000109463
ENSDART00000017155 |
npsn
|
nephrosin |
| chr19_-_25081711 | 2.40 |
ENSDART00000058513
|
xkr8.3
|
XK, Kell blood group complex subunit-related family, member 8, tandem duplicate 3 |
| chr23_+_41679586 | 2.40 |
ENSDART00000067662
|
CU914487.1
|
|
| chr17_+_8799451 | 2.40 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr23_+_28322986 | 2.40 |
ENSDART00000134710
|
birc5b
|
baculoviral IAP repeat containing 5b |
| chr15_+_45563656 | 2.40 |
ENSDART00000157501
|
cldn15lb
|
claudin 15-like b |
| chr25_+_31227747 | 2.39 |
ENSDART00000033872
|
tnni2a.1
|
troponin I type 2a (skeletal, fast), tandem duplicate 1 |
| chr2_-_2957970 | 2.39 |
ENSDART00000162505
|
si:ch1073-82l19.1
|
si:ch1073-82l19.1 |
| chr19_-_13808630 | 2.38 |
ENSDART00000166895
ENSDART00000187670 |
ctgfb
|
connective tissue growth factor b |
| chr7_-_17412559 | 2.37 |
ENSDART00000163020
|
nitr3a
|
novel immune-type receptor 3a |
| chr1_-_43727012 | 2.37 |
ENSDART00000181064
|
bdh2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr4_+_7817996 | 2.36 |
ENSDART00000166809
|
si:ch1073-67j19.1
|
si:ch1073-67j19.1 |
| chr6_+_52873822 | 2.36 |
ENSDART00000103138
|
or137-3
|
odorant receptor, family H, subfamily 137, member 3 |
| chr2_+_33326522 | 2.36 |
ENSDART00000056655
|
klf17
|
Kruppel-like factor 17 |
| chr11_-_37997419 | 2.35 |
ENSDART00000102870
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr10_-_13343831 | 2.33 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr24_-_6024466 | 2.33 |
ENSDART00000040865
|
pdss1
|
prenyl (decaprenyl) diphosphate synthase, subunit 1 |
| chr1_-_55248496 | 2.32 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr24_+_34069675 | 2.31 |
ENSDART00000143995
|
si:ch211-190p8.2
|
si:ch211-190p8.2 |
| chr7_+_2236317 | 2.30 |
ENSDART00000075859
|
zgc:172065
|
zgc:172065 |
| chr19_-_2861444 | 2.29 |
ENSDART00000169053
|
clec3bb
|
C-type lectin domain family 3, member Bb |
| chr22_+_19366866 | 2.29 |
ENSDART00000137301
|
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr24_+_19415124 | 2.29 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr12_-_5728755 | 2.28 |
ENSDART00000105887
|
dlx4b
|
distal-less homeobox 4b |
| chr23_-_19230627 | 2.28 |
ENSDART00000007122
|
guca1b
|
guanylate cyclase activator 1B |
| chr19_+_9295244 | 2.28 |
ENSDART00000132255
ENSDART00000144299 |
si:ch73-15n24.1
|
si:ch73-15n24.1 |
| chr8_-_23612462 | 2.27 |
ENSDART00000025024
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr24_-_25144441 | 2.26 |
ENSDART00000152104
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
Network of associatons between targets according to the STRING database.
First level regulatory network of si:dkey-43p13.5+uncx
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.3 | GO:0052576 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 1.8 | 5.5 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 1.8 | 9.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 1.5 | 8.8 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 1.3 | 4.0 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 1.3 | 4.0 | GO:0039015 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 1.1 | 3.3 | GO:0042730 | fibrinolysis(GO:0042730) |
| 1.1 | 4.4 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 1.0 | 5.2 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 1.0 | 4.1 | GO:0042755 | eating behavior(GO:0042755) |
| 1.0 | 2.9 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 1.0 | 3.8 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.9 | 2.8 | GO:0046379 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.9 | 45.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.9 | 2.6 | GO:0010658 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.8 | 2.4 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.8 | 2.4 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.8 | 2.3 | GO:0042546 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.8 | 2.3 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.8 | 2.3 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.7 | 2.2 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.7 | 4.2 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.7 | 13.6 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.7 | 3.4 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.7 | 2.0 | GO:0098543 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.7 | 4.6 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.6 | 1.9 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.6 | 3.1 | GO:0010872 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.6 | 2.5 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.6 | 1.8 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.6 | 1.8 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.6 | 2.9 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.6 | 2.3 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.6 | 0.6 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) |
| 0.6 | 2.3 | GO:0089709 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.6 | 3.4 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.5 | 8.4 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.5 | 2.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.5 | 3.6 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.5 | 1.5 | GO:0060262 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.5 | 3.8 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.5 | 1.4 | GO:1904251 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.5 | 9.6 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.5 | 1.4 | GO:0072592 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.4 | 1.3 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.4 | 6.2 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.4 | 1.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.4 | 1.3 | GO:0090219 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.4 | 4.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.4 | 1.2 | GO:0097435 | fibril organization(GO:0097435) |
| 0.4 | 0.4 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0014808) sarcoplasmic reticulum calcium ion transport(GO:0070296) calcium ion transport from endoplasmic reticulum to cytosol(GO:1903514) |
| 0.4 | 5.8 | GO:0061055 | myotome development(GO:0061055) |
| 0.4 | 1.5 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.4 | 1.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.4 | 2.6 | GO:1902315 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.4 | 5.5 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.3 | 5.8 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.3 | 1.7 | GO:0046083 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.3 | 3.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.3 | 2.0 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.3 | 1.3 | GO:0061033 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.3 | 1.9 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.3 | 2.8 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.3 | 2.4 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.3 | 2.4 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.3 | 1.5 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.3 | 1.2 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.3 | 2.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.3 | 14.3 | GO:0018198 | peptidyl-cysteine modification(GO:0018198) |
| 0.3 | 1.2 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.3 | 2.5 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.3 | 1.1 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.3 | 0.8 | GO:0048389 | intermediate mesoderm development(GO:0048389) |
| 0.3 | 1.9 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.3 | 2.9 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.3 | 0.8 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.3 | 1.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.3 | 3.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.3 | 2.5 | GO:0070836 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.2 | 1.0 | GO:0048308 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.2 | 0.5 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.2 | 5.3 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.2 | 5.6 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.2 | 0.9 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.2 | 8.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.2 | 3.3 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.2 | 2.6 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.2 | 5.5 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.2 | 2.4 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.2 | 1.1 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.2 | 1.5 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.2 | 1.5 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.2 | 1.7 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.2 | 2.3 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.2 | 1.9 | GO:0009204 | deoxyribonucleoside triphosphate catabolic process(GO:0009204) |
| 0.2 | 1.0 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.2 | 2.9 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.2 | 4.2 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.2 | 0.8 | GO:0048890 | epidermal cell fate specification(GO:0009957) lateral line ganglion development(GO:0048890) |
| 0.2 | 4.7 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.2 | 0.8 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.2 | 1.4 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.2 | 9.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.2 | 3.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.2 | 1.7 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.2 | 2.2 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.2 | 4.0 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.2 | 1.1 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.2 | 2.9 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.2 | 5.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.2 | 2.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 1.2 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.2 | 0.9 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.2 | 1.9 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.2 | 1.0 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.2 | 0.9 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.2 | 2.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.2 | 1.0 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.2 | 1.1 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.2 | 0.9 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 1.5 | GO:0015810 | C4-dicarboxylate transport(GO:0015740) acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.9 | GO:0060347 | trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) |
| 0.1 | 1.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 1.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 2.6 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 2.2 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 0.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 2.9 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.1 | 2.0 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 1.3 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 1.8 | GO:0007260 | tyrosine phosphorylation of STAT protein(GO:0007260) regulation of tyrosine phosphorylation of STAT protein(GO:0042509) positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 | 2.2 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 2.1 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 1.0 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 1.0 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 | 2.4 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 0.9 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.9 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.1 | 2.7 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.1 | 1.0 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.1 | 2.7 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 1.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.5 | GO:0046823 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 0.1 | 1.8 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 1.0 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.1 | 1.5 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 0.4 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.1 | 1.2 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.8 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 5.3 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 0.3 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.1 | 2.0 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.1 | 3.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 4.8 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 1.7 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 8.9 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 0.7 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.1 | 0.4 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 2.0 | GO:0032292 | peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 1.4 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 3.1 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 1.8 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 0.5 | GO:0046102 | adenosine catabolic process(GO:0006154) inosine metabolic process(GO:0046102) inosine biosynthetic process(GO:0046103) |
| 0.1 | 1.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.5 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.5 | GO:0060847 | endothelial cell fate specification(GO:0060847) |
| 0.1 | 2.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 1.1 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 0.7 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.1 | 5.3 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.1 | 0.9 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.6 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.7 | GO:0060118 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.1 | 0.7 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.1 | 0.7 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.1 | 1.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 0.3 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.1 | 0.6 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 22.7 | GO:0060537 | muscle tissue development(GO:0060537) |
| 0.1 | 1.0 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.1 | 1.0 | GO:0030819 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.1 | 0.4 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.1 | 1.6 | GO:0031936 | nucleosome positioning(GO:0016584) negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 2.6 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.1 | 1.8 | GO:0014902 | myotube differentiation(GO:0014902) |
| 0.1 | 1.0 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.1 | 9.1 | GO:0034728 | nucleosome organization(GO:0034728) |
| 0.1 | 5.0 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.1 | 1.4 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.1 | 0.4 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.1 | 1.1 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 5.5 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 0.4 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.1 | 0.8 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.7 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 2.8 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 2.1 | GO:0043434 | response to peptide hormone(GO:0043434) |
| 0.1 | 1.0 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.1 | 0.5 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 0.9 | GO:1901072 | chitin catabolic process(GO:0006032) glucosamine-containing compound catabolic process(GO:1901072) |
| 0.1 | 2.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 1.5 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.1 | 0.6 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.1 | 0.4 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.1 | 0.9 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 1.7 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 2.3 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.1 | 1.0 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 1.7 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 0.6 | GO:0008105 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.1 | 5.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 0.2 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.1 | 0.1 | GO:0070316 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 1.1 | GO:0060173 | limb development(GO:0060173) |
| 0.1 | 1.1 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.1 | 3.0 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.1 | 0.4 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.1 | 0.8 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.8 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.1 | 19.4 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.1 | 1.0 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 3.8 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 2.2 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 0.8 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 0.6 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 1.9 | GO:0006900 | membrane budding(GO:0006900) |
| 0.0 | 0.8 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 1.4 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.3 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.9 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.5 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.0 | 1.6 | GO:0030278 | regulation of ossification(GO:0030278) |
| 0.0 | 0.5 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 2.4 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 1.7 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.7 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 1.7 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.3 | GO:0002753 | cytoplasmic pattern recognition receptor signaling pathway(GO:0002753) |
| 0.0 | 0.2 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.0 | 1.9 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 1.1 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 4.9 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 2.3 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 1.0 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.8 | GO:0045785 | positive regulation of cell adhesion(GO:0045785) |
| 0.0 | 0.9 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.9 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.8 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 1.7 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 3.0 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 1.0 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.7 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.7 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.7 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.4 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.6 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.7 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 5.6 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 2.6 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.7 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.1 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.4 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.5 | GO:0019229 | regulation of vasoconstriction(GO:0019229) |
| 0.0 | 8.4 | GO:0006412 | translation(GO:0006412) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.4 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 1.2 | GO:0050871 | phagocytosis, recognition(GO:0006910) positive regulation of B cell activation(GO:0050871) |
| 0.0 | 0.9 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 1.7 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.4 | GO:0002761 | regulation of myeloid leukocyte differentiation(GO:0002761) |
| 0.0 | 0.6 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.0 | 0.3 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 1.4 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 0.2 | GO:0009583 | detection of light stimulus(GO:0009583) |
| 0.0 | 0.4 | GO:0006278 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.0 | 0.6 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 2.4 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.2 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.4 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 2.3 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.8 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 1.2 | GO:0031589 | cell-substrate adhesion(GO:0031589) |
| 0.0 | 0.1 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.3 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.1 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.0 | 0.1 | GO:0072599 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.0 | 0.2 | GO:0030282 | bone mineralization(GO:0030282) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0097189 | apoptotic body(GO:0097189) |
| 1.3 | 3.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 1.3 | 5.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.2 | 50.7 | GO:0005861 | troponin complex(GO:0005861) |
| 1.0 | 3.0 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.9 | 6.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.7 | 2.1 | GO:0019185 | snRNA-activating protein complex(GO:0019185) |
| 0.7 | 2.7 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.6 | 4.6 | GO:0071546 | pi-body(GO:0071546) |
| 0.5 | 4.0 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.5 | 6.3 | GO:0042627 | chylomicron(GO:0042627) |
| 0.4 | 4.3 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.4 | 8.9 | GO:0043186 | P granule(GO:0043186) |
| 0.3 | 2.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.3 | 0.9 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.3 | 2.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.3 | 1.1 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.3 | 3.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.3 | 16.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.3 | 0.8 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.3 | 3.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.2 | 1.0 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.2 | 5.9 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 21.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.2 | 10.2 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.2 | 0.9 | GO:0000811 | GINS complex(GO:0000811) |
| 0.2 | 0.9 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.2 | 8.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.2 | 7.8 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 1.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.2 | 2.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 2.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.2 | 0.6 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.1 | 5.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 3.6 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 1.9 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 4.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.5 | GO:0070743 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.1 | 6.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 3.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 3.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 5.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 0.5 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.1 | 1.4 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.1 | 1.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.8 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 1.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 2.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 13.5 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 0.4 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.1 | 0.4 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.1 | 1.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.9 | GO:0033290 | eukaryotic 48S preinitiation complex(GO:0033290) |
| 0.1 | 1.0 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 14.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 0.5 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 1.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.1 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 2.8 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 1.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 2.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 6.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 2.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.2 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.1 | 0.7 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 1.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 1.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 5.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 11.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 5.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 1.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 2.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 2.0 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.5 | GO:0045252 | dihydrolipoyl dehydrogenase complex(GO:0045240) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 1.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.9 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 1.2 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.8 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.4 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 1.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 9.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 3.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.8 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 36.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.7 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 2.1 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 1.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 1.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.2 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.3 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.4 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.8 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 3.6 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 7.8 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 1.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.7 | GO:0005814 | centriole(GO:0005814) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 11.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 2.2 | 11.1 | GO:0009374 | biotin binding(GO:0009374) |
| 1.5 | 7.3 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 1.3 | 5.2 | GO:0048407 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 1.1 | 3.4 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 1.1 | 6.7 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 1.1 | 3.2 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.9 | 2.8 | GO:0001635 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.9 | 2.8 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.9 | 2.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.7 | 2.9 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.7 | 2.0 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.6 | 3.1 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.6 | 3.5 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.6 | 1.7 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.6 | 1.7 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.6 | 1.7 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.5 | 3.8 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.5 | 4.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.5 | 3.6 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.5 | 3.8 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.5 | 2.8 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.5 | 1.4 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
| 0.4 | 2.2 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.4 | 1.8 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.4 | 2.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.4 | 5.6 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.4 | 3.4 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.4 | 2.6 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.4 | 1.5 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.4 | 2.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.4 | 2.5 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.4 | 6.1 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.3 | 5.8 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.3 | 1.7 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.3 | 1.9 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.3 | 9.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.3 | 6.4 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.3 | 2.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.3 | 4.5 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.3 | 6.6 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.3 | 2.0 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.3 | 2.3 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.3 | 3.1 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.3 | 5.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.3 | 3.3 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.3 | 0.8 | GO:0000035 | acyl binding(GO:0000035) |
| 0.3 | 1.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.3 | 2.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.3 | 3.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.3 | 0.8 | GO:0032138 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.3 | 3.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.3 | 1.8 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.2 | 7.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.2 | 1.0 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.2 | 4.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 2.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 1.3 | GO:0016793 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) |
| 0.2 | 6.4 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.2 | 1.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.2 | 3.7 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 1.5 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.2 | 2.3 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.2 | 1.0 | GO:0015140 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.2 | 1.0 | GO:0098634 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.2 | 9.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.2 | 0.6 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.2 | 1.7 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.2 | 2.4 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.2 | 2.9 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 7.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.2 | 0.9 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 1.9 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.2 | 1.4 | GO:0015157 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 2.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.2 | 2.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 30.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.2 | 1.5 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.2 | 0.9 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.2 | 10.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.2 | 0.8 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.2 | 1.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 2.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 3.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 4.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.5 | GO:0019972 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.1 | 1.8 | GO:0008061 | chitin binding(GO:0008061) |
| 0.1 | 1.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.5 | GO:0004945 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 1.5 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 2.7 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 1.6 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 0.6 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 1.6 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.4 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.1 | 1.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 2.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.9 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 2.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.6 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 1.3 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 1.1 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.4 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 3.0 | GO:0099589 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.1 | 0.5 | GO:0001948 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.1 | 1.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.6 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.6 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 2.1 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.1 | 0.3 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.1 | 1.0 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 1.9 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 5.4 | GO:0048029 | monosaccharide binding(GO:0048029) |
| 0.1 | 2.9 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.4 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 0.9 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.1 | 0.8 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.4 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 1.0 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 10.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.8 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 0.7 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.1 | 1.0 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 1.5 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 2.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 1.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 2.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 1.6 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 1.5 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 2.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 3.2 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 4.4 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.1 | 0.7 | GO:0051119 | sugar transmembrane transporter activity(GO:0051119) |
| 0.1 | 1.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.6 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.3 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.1 | 0.9 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 2.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 1.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 14.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.5 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 4.8 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 0.8 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 4.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.9 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.2 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 3.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.3 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 6.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 2.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 1.4 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 0.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 1.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 3.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 2.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 0.8 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 7.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 2.6 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 5.7 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.8 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 3.1 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 7.3 | GO:0004175 | endopeptidase activity(GO:0004175) |
| 0.0 | 0.2 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.0 | 0.5 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.0 | 0.9 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 1.0 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 9.3 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 1.0 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.3 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.2 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.6 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 1.3 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 1.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 2.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.9 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 2.1 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 1.3 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 12.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 1.1 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 1.7 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 1.6 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.8 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 5.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 2.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 2.2 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 4.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 1.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.8 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 1.3 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.6 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.7 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 2.8 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 2.8 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.6 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.8 | GO:0005254 | chloride channel activity(GO:0005254) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.5 | 6.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.3 | 5.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 2.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 2.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.2 | 6.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 14.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.2 | 4.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.2 | 3.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 1.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 1.9 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 4.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 1.5 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 2.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 2.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 2.0 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 0.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.2 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 2.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 0.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 2.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 3.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 10.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 10.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 0.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 0.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 1.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.8 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 2.0 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.7 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 4.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 8.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.6 | 5.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.5 | 13.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.5 | 5.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.5 | 3.0 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.5 | 3.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.5 | 1.4 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.4 | 4.0 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.4 | 2.9 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.4 | 2.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.3 | 1.9 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.3 | 8.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 30.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.3 | 2.8 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.2 | 4.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 3.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 10.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 1.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.2 | 4.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 6.7 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.1 | 3.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 2.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 0.9 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 2.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 3.1 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 1.7 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 0.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 0.9 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 2.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 0.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 5.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 3.0 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 0.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 2.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 0.9 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 1.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 0.9 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.1 | 2.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 2.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 2.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 0.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.1 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 2.2 | REACTOME CHROMOSOME MAINTENANCE | Genes involved in Chromosome Maintenance |
| 0.0 | 1.2 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.5 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.1 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 1.0 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.6 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 1.4 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.2 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |