Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
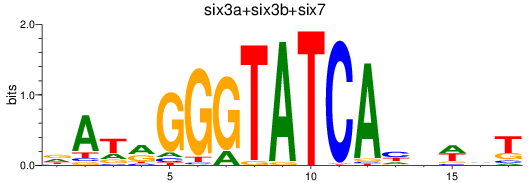
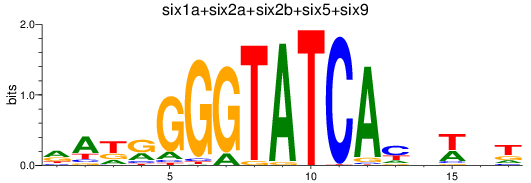
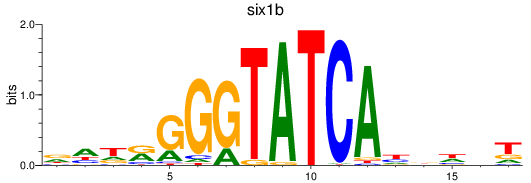
Results for six3a+six3b+six7_six1a+six2a+six2b+six5+six9_six1b
Z-value: 0.46
Transcription factors associated with six3a+six3b+six7_six1a+six2a+six2b+six5+six9_six1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
six3b
|
ENSDARG00000054879 | SIX homeobox 3b |
|
six3a
|
ENSDARG00000058008 | SIX homeobox 3a |
|
six7
|
ENSDARG00000070107 | SIX homeobox 7 |
|
six3a
|
ENSDARG00000114971 | SIX homeobox 3a |
|
six1a
|
ENSDARG00000039304 | SIX homeobox 1a |
|
six2b
|
ENSDARG00000054878 | SIX homeobox 2b |
|
six2a
|
ENSDARG00000058004 | SIX homeobox 2a |
|
six5
|
ENSDARG00000068406 | SIX homeobox 5 |
|
six9
|
ENSDARG00000068407 | SIX homeobox 9 |
|
six2a
|
ENSDARG00000115214 | SIX homeobox 2a |
|
six1b
|
ENSDARG00000026473 | SIX homeobox 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| six1a | dr11_v1_chr13_-_31622195_31622195 | -0.52 | 1.1e-07 | Click! |
| six2b | dr11_v1_chr12_-_25612170_25612170 | -0.50 | 3.2e-07 | Click! |
| six2a | dr11_v1_chr13_+_10232695_10232695 | -0.50 | 3.7e-07 | Click! |
| six1b | dr11_v1_chr20_+_20499869_20499869 | -0.48 | 1.1e-06 | Click! |
| six3a | dr11_v1_chr13_-_10261383_10261383 | -0.18 | 7.7e-02 | Click! |
| six3b | dr11_v1_chr12_+_25600685_25600685 | -0.12 | 2.5e-01 | Click! |
| six7 | dr11_v1_chr7_-_7420301_7420301 | -0.11 | 2.7e-01 | Click! |
| six9 | dr11_v1_chr18_-_37241080_37241115 | -0.10 | 3.5e-01 | Click! |
| six5 | dr11_v1_chr18_-_37252036_37252036 | -0.10 | 3.6e-01 | Click! |
Activity profile of six3a+six3b+six7_six1a+six2a+six2b+six5+six9_six1b motif
Sorted Z-values of six3a+six3b+six7_six1a+six2a+six2b+six5+six9_six1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_49283058 | 4.28 |
ENSDART00000076554
|
si:zfos-464b6.2
|
si:zfos-464b6.2 |
| chr3_-_49815223 | 2.98 |
ENSDART00000181208
|
gcgra
|
glucagon receptor a |
| chr7_-_71486162 | 2.47 |
ENSDART00000045253
|
aco1
|
aconitase 1, soluble |
| chr5_+_9377005 | 1.96 |
ENSDART00000124924
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr10_+_42898103 | 1.77 |
ENSDART00000015872
|
zcchc9
|
zinc finger, CCHC domain containing 9 |
| chr11_-_42750626 | 1.68 |
ENSDART00000130640
|
si:ch73-106k19.5
|
si:ch73-106k19.5 |
| chr8_+_20440297 | 1.64 |
ENSDART00000189486
|
mknk2b
|
MAP kinase interacting serine/threonine kinase 2b |
| chr16_+_41060161 | 1.30 |
ENSDART00000141130
|
scap
|
SREBF chaperone |
| chr16_-_46567344 | 1.27 |
ENSDART00000127721
|
si:dkey-152b24.7
|
si:dkey-152b24.7 |
| chr17_+_16429826 | 1.20 |
ENSDART00000136078
|
efcab11
|
EF-hand calcium binding domain 11 |
| chr3_-_21094437 | 1.16 |
ENSDART00000153739
ENSDART00000109790 |
nlk1
|
nemo-like kinase, type 1 |
| chr21_+_11385031 | 1.09 |
ENSDART00000040598
|
cel.2
|
carboxyl ester lipase, tandem duplicate 2 |
| chr21_-_26028205 | 1.07 |
ENSDART00000034875
|
sdf2
|
stromal cell-derived factor 2 |
| chr8_-_26961779 | 1.04 |
ENSDART00000099214
|
slc16a1b
|
solute carrier family 16 (monocarboxylate transporter), member 1b |
| chr23_+_39481537 | 1.03 |
ENSDART00000187222
|
mical1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr22_-_24967348 | 0.99 |
ENSDART00000153490
ENSDART00000084871 |
fam20cl
|
family with sequence similarity 20, member C like |
| chr14_-_38872536 | 0.98 |
ENSDART00000184315
ENSDART00000127479 |
gsr
|
glutathione reductase |
| chr14_+_22397251 | 0.97 |
ENSDART00000185239
ENSDART00000124072 ENSDART00000054977 |
atp7a
|
ATPase copper transporting alpha |
| chr21_-_27195256 | 0.89 |
ENSDART00000133152
ENSDART00000065401 |
zgc:110782
|
zgc:110782 |
| chr11_-_44194132 | 0.85 |
ENSDART00000182954
ENSDART00000111271 |
CABZ01080074.1
|
|
| chr14_-_38873095 | 0.84 |
ENSDART00000047050
ENSDART00000173285 ENSDART00000147521 |
gsr
|
glutathione reductase |
| chr11_-_41220794 | 0.78 |
ENSDART00000192895
|
mrps16
|
mitochondrial ribosomal protein S16 |
| chr12_+_13742953 | 0.76 |
ENSDART00000152257
|
ppp1r16a
|
protein phosphatase 1, regulatory subunit 16A |
| chr9_-_18911608 | 0.74 |
ENSDART00000138785
|
si:dkey-239h2.3
|
si:dkey-239h2.3 |
| chr12_+_13742778 | 0.71 |
ENSDART00000111401
ENSDART00000190552 |
ppp1r16a
|
protein phosphatase 1, regulatory subunit 16A |
| chr21_-_7940043 | 0.67 |
ENSDART00000099733
ENSDART00000136671 |
f2rl1.1
|
coagulation factor II (thrombin) receptor-like 1, tandem duplicate 1 |
| chr19_+_34230108 | 0.65 |
ENSDART00000141950
|
galnt12
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 |
| chr1_-_55896332 | 0.64 |
ENSDART00000147516
|
si:ch73-343l4.8
|
si:ch73-343l4.8 |
| chr7_-_38183331 | 0.61 |
ENSDART00000149382
|
abcc12
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 12 |
| chr10_+_35468978 | 0.61 |
ENSDART00000131984
|
phldb2a
|
pleckstrin homology-like domain, family B, member 2a |
| chr11_-_24016761 | 0.59 |
ENSDART00000153601
ENSDART00000067817 ENSDART00000170531 |
chia.3
|
chitinase, acidic.3 |
| chr19_-_20403845 | 0.56 |
ENSDART00000151265
ENSDART00000147911 ENSDART00000151356 |
dazl
|
deleted in azoospermia-like |
| chr20_-_45709990 | 0.56 |
ENSDART00000027482
|
gpcpd1
|
glycerophosphocholine phosphodiesterase 1 |
| chr1_-_24255149 | 0.55 |
ENSDART00000146960
|
lrba
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr13_+_9678427 | 0.55 |
ENSDART00000153907
|
sema4ga
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ga |
| chr24_-_36175365 | 0.51 |
ENSDART00000065338
|
pak1ip1
|
PAK1 interacting protein 1 |
| chr1_-_21723329 | 0.51 |
ENSDART00000137138
|
si:ch211-134c9.2
|
si:ch211-134c9.2 |
| chr2_+_49631850 | 0.51 |
ENSDART00000114274
ENSDART00000114516 |
si:dkey-53k12.18
|
si:dkey-53k12.18 |
| chr2_-_51512294 | 0.50 |
ENSDART00000185287
|
si:ch211-9d9.8
|
si:ch211-9d9.8 |
| chr25_-_21763750 | 0.48 |
ENSDART00000089596
|
tmem168b
|
transmembrane protein 168b |
| chr15_+_41815703 | 0.46 |
ENSDART00000059508
|
pxylp1
|
2-phosphoxylose phosphatase 1 |
| chr4_+_74396786 | 0.46 |
ENSDART00000127501
ENSDART00000174347 |
zmp:0000001020
|
zmp:0000001020 |
| chr3_+_1724941 | 0.45 |
ENSDART00000193402
|
BX321875.2
|
|
| chr4_+_73672430 | 0.42 |
ENSDART00000174310
ENSDART00000150505 |
si:dkey-262g12.7
|
si:dkey-262g12.7 |
| chr13_+_9678262 | 0.42 |
ENSDART00000111365
|
sema4ga
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ga |
| chr5_+_36900157 | 0.40 |
ENSDART00000183533
ENSDART00000051184 |
hnrnpl
|
heterogeneous nuclear ribonucleoprotein L |
| chr8_+_14381272 | 0.38 |
ENSDART00000057642
|
acbd6
|
acyl-CoA binding domain containing 6 |
| chr7_-_35708450 | 0.37 |
ENSDART00000193886
|
irx5a
|
iroquois homeobox 5a |
| chr5_+_36899691 | 0.35 |
ENSDART00000132322
|
hnrnpl
|
heterogeneous nuclear ribonucleoprotein L |
| chr20_-_39735952 | 0.35 |
ENSDART00000101049
ENSDART00000137485 ENSDART00000062402 |
tpd52l1
|
tumor protein D52-like 1 |
| chr10_+_14488625 | 0.34 |
ENSDART00000026383
|
sigmar1
|
sigma non-opioid intracellular receptor 1 |
| chr5_-_54395488 | 0.33 |
ENSDART00000160781
|
zmynd19
|
zinc finger, MYND-type containing 19 |
| chr16_-_23797570 | 0.33 |
ENSDART00000077834
|
rps27.2
|
ribosomal protein S27, isoform 2 |
| chr14_-_8814590 | 0.32 |
ENSDART00000106670
|
trmt112
|
tRNA methyltransferase 11-2 homolog (S. cerevisiae) |
| chr20_+_13783040 | 0.32 |
ENSDART00000115329
ENSDART00000152497 |
lpgat1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr17_+_132555 | 0.32 |
ENSDART00000158159
|
zgc:77287
|
zgc:77287 |
| chr4_+_18782539 | 0.31 |
ENSDART00000101052
|
slc26a4
|
solute carrier family 26 (anion exchanger), member 4 |
| chr4_+_18782840 | 0.30 |
ENSDART00000146450
|
slc26a4
|
solute carrier family 26 (anion exchanger), member 4 |
| chr2_-_24270062 | 0.30 |
ENSDART00000192445
|
myh7
|
myosin heavy chain 7 |
| chr10_+_23520952 | 0.30 |
ENSDART00000125317
|
zmp:0000000937
|
zmp:0000000937 |
| chr2_+_6999369 | 0.30 |
ENSDART00000050597
|
xpr1b
|
xenotropic and polytropic retrovirus receptor 1b |
| chr25_+_3788074 | 0.30 |
ENSDART00000154008
|
chid1
|
chitinase domain containing 1 |
| chr13_+_33368140 | 0.30 |
ENSDART00000033848
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr4_-_5317006 | 0.29 |
ENSDART00000150867
|
si:ch211-214j24.15
|
si:ch211-214j24.15 |
| chr6_-_40352215 | 0.28 |
ENSDART00000103992
|
ttll3
|
tubulin tyrosine ligase-like family, member 3 |
| chr17_+_13099476 | 0.25 |
ENSDART00000012670
|
pnn
|
pinin, desmosome associated protein |
| chr10_+_40633990 | 0.22 |
ENSDART00000190489
ENSDART00000139474 |
si:ch211-238p8.31
|
si:ch211-238p8.31 |
| chr18_+_33322515 | 0.22 |
ENSDART00000136603
|
v2ra18
|
vomeronasal 2 receptor, a18 |
| chr16_-_11961325 | 0.22 |
ENSDART00000143845
|
si:ch73-296e2.3
|
si:ch73-296e2.3 |
| chr11_+_24002503 | 0.22 |
ENSDART00000164702
|
chia.2
|
chitinase, acidic.2 |
| chr5_-_26247973 | 0.20 |
ENSDART00000098527
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr25_+_192116 | 0.20 |
ENSDART00000153983
|
zgc:114188
|
zgc:114188 |
| chr16_-_46567136 | 0.20 |
ENSDART00000159180
|
si:dkey-152b24.7
|
si:dkey-152b24.7 |
| chr21_-_21465111 | 0.19 |
ENSDART00000141487
|
nectin3b
|
nectin cell adhesion molecule 3b |
| chr6_+_41181869 | 0.19 |
ENSDART00000002046
|
opn1mw1
|
opsin 1 (cone pigments), medium-wave-sensitive, 1 |
| chr13_-_3324764 | 0.19 |
ENSDART00000102748
ENSDART00000114040 |
ubr2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr11_+_24339377 | 0.18 |
ENSDART00000133679
ENSDART00000135435 ENSDART00000017973 ENSDART00000131365 ENSDART00000186418 |
rbm39a
|
RNA binding motif protein 39a |
| chr20_+_34502606 | 0.18 |
ENSDART00000139739
|
gorab
|
golgin, rab6-interacting |
| chr13_-_18122333 | 0.18 |
ENSDART00000128748
|
washc2c
|
WASH complex subunit 2C |
| chr20_+_46237000 | 0.17 |
ENSDART00000125636
|
taar14a
|
trace amine associated receptor 14a |
| chr11_+_3575963 | 0.17 |
ENSDART00000077305
|
si:dkey-33m11.8
|
si:dkey-33m11.8 |
| chr19_-_11949996 | 0.16 |
ENSDART00000163478
ENSDART00000167299 |
cpsf1
|
cleavage and polyadenylation specific factor 1 |
| chr21_-_13972745 | 0.16 |
ENSDART00000143874
|
akna
|
AT-hook transcription factor |
| chr4_-_13502549 | 0.16 |
ENSDART00000140366
|
si:ch211-266a5.12
|
si:ch211-266a5.12 |
| chr5_+_13326765 | 0.15 |
ENSDART00000090851
|
ydjc
|
YdjC chitooligosaccharide deacetylase homolog |
| chr24_-_12745222 | 0.15 |
ENSDART00000151836
|
si:ch211-196f5.9
|
si:ch211-196f5.9 |
| chr2_-_36494308 | 0.14 |
ENSDART00000110378
|
BX901889.2
|
|
| chr19_+_10485847 | 0.14 |
ENSDART00000135719
|
si:ch211-171h4.6
|
si:ch211-171h4.6 |
| chr20_-_34127415 | 0.14 |
ENSDART00000010028
|
ptgs2b
|
prostaglandin-endoperoxide synthase 2b |
| chr8_+_7801060 | 0.13 |
ENSDART00000161618
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr11_+_1602916 | 0.13 |
ENSDART00000184434
ENSDART00000112597 ENSDART00000192165 |
si:dkey-40c23.2
si:dkey-40c23.3
|
si:dkey-40c23.2 si:dkey-40c23.3 |
| chr24_+_30435164 | 0.13 |
ENSDART00000193877
|
dpyda.1
|
dihydropyrimidine dehydrogenase a, tandem duplicate 1 |
| chr12_-_35095414 | 0.12 |
ENSDART00000153229
|
si:dkey-21e13.3
|
si:dkey-21e13.3 |
| chr5_+_41477526 | 0.12 |
ENSDART00000153567
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr10_-_32610776 | 0.12 |
ENSDART00000017436
|
mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr8_+_44478294 | 0.12 |
ENSDART00000006898
|
si:ch73-211l2.3
|
si:ch73-211l2.3 |
| chr15_-_38218320 | 0.11 |
ENSDART00000152363
|
rhoga
|
ras homolog family member Ga |
| chr17_-_6600899 | 0.11 |
ENSDART00000154074
ENSDART00000180912 |
ANKRD66
|
si:ch211-189e2.2 |
| chr6_-_18618106 | 0.11 |
ENSDART00000161562
|
znf207b
|
zinc finger protein 207, b |
| chr6_-_1514767 | 0.11 |
ENSDART00000067586
|
chchd6b
|
coiled-coil-helix-coiled-coil-helix domain containing 6b |
| chr13_+_25549425 | 0.10 |
ENSDART00000087553
ENSDART00000169199 |
sec23ip
|
SEC23 interacting protein |
| chr4_-_49720388 | 0.09 |
ENSDART00000154146
|
znf1057
|
zinc finger protein 1057 |
| chr20_-_44460789 | 0.09 |
ENSDART00000085442
|
mut
|
methylmalonyl CoA mutase |
| chr14_-_1355544 | 0.09 |
ENSDART00000060417
|
cetn4
|
centrin 4 |
| chr4_-_5332814 | 0.09 |
ENSDART00000144225
|
osgep
|
O-sialoglycoprotein endopeptidase |
| chr14_+_1355857 | 0.09 |
ENSDART00000188008
|
bbs12
|
Bardet-Biedl syndrome 12 |
| chr6_+_52947699 | 0.08 |
ENSDART00000180913
|
uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr19_-_22664128 | 0.08 |
ENSDART00000136295
|
eif3eb
|
eukaryotic translation initiation factor 3, subunit E, b |
| chr12_+_28854963 | 0.08 |
ENSDART00000153227
|
nfe2l1b
|
nuclear factor, erythroid 2-like 1b |
| chr15_-_945804 | 0.08 |
ENSDART00000063257
|
alox5b.1
|
arachidonate 5-lipoxygenase b, tandem duplicate 1 |
| chr9_-_28990649 | 0.08 |
ENSDART00000078823
|
ptpn4a
|
protein tyrosine phosphatase, non-receptor type 4a |
| chr16_+_14201401 | 0.08 |
ENSDART00000113679
|
dap3
|
death associated protein 3 |
| chr16_-_383664 | 0.07 |
ENSDART00000051693
|
irx4a
|
iroquois homeobox 4a |
| chr15_+_37546391 | 0.07 |
ENSDART00000186625
|
psenen
|
presenilin enhancer gamma secretase subunit |
| chr23_+_3721042 | 0.07 |
ENSDART00000143323
|
smim29
|
small integral membrane protein 29 |
| chr7_-_40657831 | 0.07 |
ENSDART00000084153
|
nom1
|
nucleolar protein with MIF4G domain 1 |
| chr5_-_41841892 | 0.06 |
ENSDART00000167089
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr2_-_51275873 | 0.06 |
ENSDART00000168019
|
si:ch211-215e19.4
|
si:ch211-215e19.4 |
| chr4_-_5538752 | 0.06 |
ENSDART00000180219
|
BX324164.5
|
|
| chr7_-_59515569 | 0.06 |
ENSDART00000163343
ENSDART00000165457 ENSDART00000163745 |
slx1b
|
SLX1 homolog B, structure-specific endonuclease subunit |
| chr17_+_4408809 | 0.06 |
ENSDART00000113401
|
si:zfos-364h11.1
|
si:zfos-364h11.1 |
| chr7_-_29356084 | 0.05 |
ENSDART00000075757
|
gtf2a2
|
general transcription factor IIA, 2 |
| chr25_+_19008497 | 0.05 |
ENSDART00000104420
|
samm50
|
SAMM50 sorting and assembly machinery component |
| chr18_+_33284202 | 0.05 |
ENSDART00000132758
|
v2ra17
|
vomeronasal 2 receptor, a17 |
| chr21_+_18405585 | 0.04 |
ENSDART00000139318
|
si:dkey-1d7.3
|
si:dkey-1d7.3 |
| chr12_+_19030391 | 0.04 |
ENSDART00000153927
|
si:ch73-139e5.2
|
si:ch73-139e5.2 |
| chr22_-_5171362 | 0.04 |
ENSDART00000124889
|
tnfaip8l1
|
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr8_+_23745167 | 0.03 |
ENSDART00000149352
|
si:ch211-163l21.11
|
si:ch211-163l21.11 |
| chr25_-_37180969 | 0.03 |
ENSDART00000152338
|
tdrd12
|
tudor domain containing 12 |
| chr5_+_41477954 | 0.02 |
ENSDART00000185871
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr20_+_4222357 | 0.01 |
ENSDART00000188331
|
ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr22_+_2315996 | 0.01 |
ENSDART00000132489
|
znf1175
|
zinc finger protein 1175 |
| chr9_+_53637932 | 0.01 |
ENSDART00000188962
|
CABZ01084081.1
|
|
| chr15_-_15910226 | 0.00 |
ENSDART00000154219
|
synrg
|
synergin, gamma |
| chr2_-_38312757 | 0.00 |
ENSDART00000167274
|
ap1g2
|
adaptor-related protein complex 1, gamma 2 subunit |
| chr4_+_20085114 | 0.00 |
ENSDART00000186698
ENSDART00000188635 |
ppp6r2a
|
protein phosphatase 6, regulatory subunit 2a |
Network of associatons between targets according to the STRING database.
First level regulatory network of six3a+six3b+six7_six1a+six2a+six2b+six5+six9_six1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.5 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.3 | 1.0 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.2 | 1.0 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.2 | 1.0 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 1.3 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 1.1 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.6 | GO:0060965 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.1 | 0.5 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.1 | 0.7 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.3 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.1 | 1.8 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 0.3 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 0.8 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.1 | 1.8 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 1.1 | GO:0071712 | protein O-linked mannosylation(GO:0035269) ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 3.0 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.1 | 0.3 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.6 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.0 | 1.5 | GO:0035304 | regulation of protein dephosphorylation(GO:0035304) |
| 0.0 | 0.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.3 | GO:0030320 | microglia differentiation(GO:0014004) cellular anion homeostasis(GO:0030002) cellular monovalent inorganic anion homeostasis(GO:0030320) cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.4 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 1.2 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0006212 | uracil catabolic process(GO:0006212) uracil metabolic process(GO:0019860) |
| 0.0 | 0.6 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.2 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.4 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.0 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.3 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.1 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 1.0 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.2 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.3 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.6 | 3.0 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.4 | 1.5 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.4 | 1.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.4 | 1.8 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.2 | 1.0 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.2 | 0.6 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.1 | 0.3 | GO:0015562 | inositol hexakisphosphate binding(GO:0000822) efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.3 | GO:0070735 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.1 | 1.0 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.7 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 1.1 | GO:0008061 | chitin binding(GO:0008061) |
| 0.1 | 1.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.6 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.3 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 1.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 2.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 1.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.1 | GO:0017113 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.0 | 1.3 | GO:0032934 | sterol binding(GO:0032934) |
| 0.0 | 1.0 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 4.9 | GO:0016757 | transferase activity, transferring glycosyl groups(GO:0016757) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 1.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |