Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
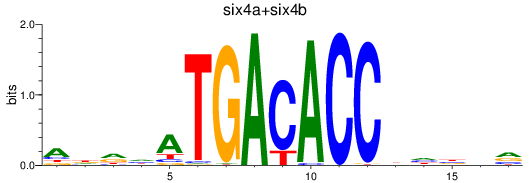
Results for six4a+six4b
Z-value: 0.73
Transcription factors associated with six4a+six4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
six4a
|
ENSDARG00000004695 | SIX homeobox 4a |
|
six4b
|
ENSDARG00000031983 | SIX homeobox 4b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| six4b | dr11_v1_chr20_+_20484827_20484827 | -0.26 | 1.3e-02 | Click! |
| six4a | dr11_v1_chr13_-_31647323_31647323 | -0.13 | 2.0e-01 | Click! |
Activity profile of six4a+six4b motif
Sorted Z-values of six4a+six4b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_10134345 | 8.61 |
ENSDART00000100725
|
ahsg2
|
alpha-2-HS-glycoprotein 2 |
| chr9_+_8396755 | 7.48 |
ENSDART00000043067
|
zgc:171776
|
zgc:171776 |
| chr3_-_39152478 | 6.91 |
ENSDART00000154550
|
si:dkeyp-57f11.2
|
si:dkeyp-57f11.2 |
| chr2_+_24304854 | 5.60 |
ENSDART00000078972
|
fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr22_+_25236888 | 5.23 |
ENSDART00000037286
|
zgc:172218
|
zgc:172218 |
| chr22_+_25242322 | 5.10 |
ENSDART00000134628
|
si:ch211-226h8.8
|
si:ch211-226h8.8 |
| chr1_+_55140970 | 5.08 |
ENSDART00000039807
|
mb
|
myoglobin |
| chr24_+_34089977 | 5.05 |
ENSDART00000157466
|
asb10
|
ankyrin repeat and SOCS box containing 10 |
| chr22_+_25236657 | 4.64 |
ENSDART00000138012
|
zgc:172218
|
zgc:172218 |
| chr14_-_7128980 | 4.22 |
ENSDART00000171311
|
si:ch73-43g23.1
|
si:ch73-43g23.1 |
| chr9_+_2041535 | 3.81 |
ENSDART00000093187
|
lnpa
|
limb and neural patterns a |
| chr15_-_35126332 | 3.27 |
ENSDART00000007636
|
zgc:55413
|
zgc:55413 |
| chr8_+_35964482 | 3.10 |
ENSDART00000129357
ENSDART00000154953 |
glt1d1
|
glycosyltransferase 1 domain containing 1 |
| chr5_-_5831037 | 2.86 |
ENSDART00000112856
|
zmp:0000000846
|
zmp:0000000846 |
| chr16_-_13818061 | 2.63 |
ENSDART00000132982
ENSDART00000144856 |
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr7_-_41851605 | 2.61 |
ENSDART00000142981
|
mylk3
|
myosin light chain kinase 3 |
| chr25_-_15214161 | 2.46 |
ENSDART00000031499
|
wt1a
|
wilms tumor 1a |
| chr8_+_49149907 | 2.31 |
ENSDART00000138746
|
snpha
|
syntaphilin a |
| chr3_-_56896702 | 2.31 |
ENSDART00000023265
|
ush1ga
|
Usher syndrome 1Ga (autosomal recessive) |
| chr6_-_3978919 | 2.18 |
ENSDART00000167753
|
slc25a12
|
solute carrier family 25 (aspartate/glutamate carrier), member 12 |
| chr22_-_36774057 | 2.17 |
ENSDART00000125048
|
acy1
|
aminoacylase 1 |
| chr20_+_33534038 | 2.12 |
ENSDART00000029206
|
kcnf1a
|
potassium voltage-gated channel, subfamily F, member 1a |
| chr19_-_10330778 | 2.09 |
ENSDART00000081465
ENSDART00000136653 ENSDART00000171232 |
ccdc106b
|
coiled-coil domain containing 106b |
| chr23_+_45785563 | 2.05 |
ENSDART00000186027
|
CABZ01088036.1
|
|
| chr7_+_6480212 | 2.04 |
ENSDART00000173320
|
si:cabz01036022.1
|
si:cabz01036022.1 |
| chr11_+_41560792 | 2.01 |
ENSDART00000127292
|
kcnab2a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 a |
| chr21_+_34167178 | 2.00 |
ENSDART00000158308
|
trpc5b
|
transient receptor potential cation channel, subfamily C, member 5b |
| chr5_+_55221593 | 1.85 |
ENSDART00000073638
|
tmc2a
|
transmembrane channel-like 2a |
| chr1_-_7894255 | 1.85 |
ENSDART00000167126
ENSDART00000145460 |
radil
|
Ras association and DIL domains |
| chr12_-_18872927 | 1.82 |
ENSDART00000187717
|
shisa8b
|
shisa family member 8b |
| chr17_-_1407593 | 1.82 |
ENSDART00000157622
ENSDART00000159458 |
zbtb42
|
zinc finger and BTB domain containing 42 |
| chr22_-_36774219 | 1.82 |
ENSDART00000056151
ENSDART00000168711 |
acy1
|
aminoacylase 1 |
| chr6_-_33129312 | 1.78 |
ENSDART00000156987
|
tspan1
|
tetraspanin 1 |
| chr13_-_39399967 | 1.78 |
ENSDART00000190791
ENSDART00000136267 |
slc35f3b
|
solute carrier family 35, member F3b |
| chr6_+_40591149 | 1.71 |
ENSDART00000189060
ENSDART00000188298 |
frs3
|
fibroblast growth factor receptor substrate 3 |
| chr11_+_583725 | 1.69 |
ENSDART00000189415
|
mkrn2os.2
|
MKRN2 opposite strand, tandem duplicate 2 |
| chr22_-_209741 | 1.69 |
ENSDART00000171954
|
ora1
|
olfactory receptor class A related 1 |
| chr2_+_29491314 | 1.67 |
ENSDART00000181774
|
dlgap1a
|
discs, large (Drosophila) homolog-associated protein 1a |
| chr3_+_12718100 | 1.66 |
ENSDART00000162343
ENSDART00000192425 |
cyp2k20
|
cytochrome P450, family 2, subfamily k, polypeptide 20 |
| chr22_+_15310103 | 1.65 |
ENSDART00000145849
|
CU104797.1
|
|
| chr19_+_770300 | 1.63 |
ENSDART00000062518
|
gstr
|
glutathione S-transferase rho |
| chr10_-_32610776 | 1.61 |
ENSDART00000017436
|
mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr19_+_31183495 | 1.58 |
ENSDART00000088618
|
meox2b
|
mesenchyme homeobox 2b |
| chr6_-_21678263 | 1.58 |
ENSDART00000038777
|
si:dkey-43k4.5
|
si:dkey-43k4.5 |
| chr1_-_58561963 | 1.55 |
ENSDART00000165040
|
slc27a1b
|
solute carrier family 27 (fatty acid transporter), member 1b |
| chr6_+_39085969 | 1.53 |
ENSDART00000004240
|
prss60.1
|
protease, serine, 60.1 |
| chr2_+_38804223 | 1.52 |
ENSDART00000147939
|
carmil3
|
capping protein regulator and myosin 1 linker 3 |
| chr23_+_16469530 | 1.52 |
ENSDART00000132898
|
ntsr1
|
neurotensin receptor 1 (high affinity) |
| chr3_-_34072446 | 1.51 |
ENSDART00000151528
|
ighv8-2
|
immunoglobulin heavy variable 8-2 |
| chr2_+_25198648 | 1.49 |
ENSDART00000110922
|
ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr17_+_5768608 | 1.47 |
ENSDART00000157039
|
rp1l1a
|
retinitis pigmentosa 1-like 1a |
| chr11_+_40649412 | 1.47 |
ENSDART00000043016
ENSDART00000134560 |
slc45a1
|
solute carrier family 45, member 1 |
| chr15_-_37834433 | 1.46 |
ENSDART00000189748
|
si:dkey-238d18.3
|
si:dkey-238d18.3 |
| chr3_+_39099716 | 1.44 |
ENSDART00000083388
|
tmem98
|
transmembrane protein 98 |
| chr21_+_21679086 | 1.44 |
ENSDART00000146225
|
or125-5
|
odorant receptor, family E, subfamily 125, member 5 |
| chr7_-_26306546 | 1.43 |
ENSDART00000140817
|
zgc:77439
|
zgc:77439 |
| chr8_+_49570884 | 1.41 |
ENSDART00000182117
ENSDART00000108613 |
rasef
|
RAS and EF-hand domain containing |
| chr8_+_14058646 | 1.37 |
ENSDART00000080852
|
ugt5e1
|
UDP glucuronosyltransferase 5 family, polypeptide E1 |
| chr2_-_32558795 | 1.36 |
ENSDART00000140026
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr23_-_7125494 | 1.36 |
ENSDART00000111929
|
slco4a1
|
solute carrier organic anion transporter family, member 4A1 |
| chr3_+_311833 | 1.35 |
ENSDART00000187375
|
FP326649.2
|
|
| chr14_-_2209742 | 1.31 |
ENSDART00000054889
|
pcdh2ab5
|
protocadherin 2 alpha b 5 |
| chr19_-_18626515 | 1.29 |
ENSDART00000160624
|
rps18
|
ribosomal protein S18 |
| chr23_-_10831995 | 1.26 |
ENSDART00000142533
|
pdzrn3a
|
PDZ domain containing RING finger 3a |
| chr4_-_77332032 | 1.24 |
ENSDART00000168628
ENSDART00000172025 |
slco1f3
|
solute carrier organic anion transporter family, member 1F3 |
| chr17_-_42213285 | 1.23 |
ENSDART00000140549
|
nkx2.2a
|
NK2 homeobox 2a |
| chr12_-_16524727 | 1.21 |
ENSDART00000166645
|
FO704724.1
|
|
| chr4_-_77506362 | 1.19 |
ENSDART00000174387
ENSDART00000181181 |
CABZ01087415.1
|
|
| chr4_-_72468168 | 1.15 |
ENSDART00000182995
ENSDART00000174067 |
CR788316.1
|
|
| chr7_+_73827805 | 1.12 |
ENSDART00000109316
|
zgc:173587
|
zgc:173587 |
| chr7_-_73852594 | 1.09 |
ENSDART00000183194
|
zgc:165555
|
zgc:165555 |
| chr1_-_7603734 | 1.08 |
ENSDART00000009315
|
mxb
|
myxovirus (influenza) resistance B |
| chr10_-_36682509 | 1.07 |
ENSDART00000148093
ENSDART00000063365 |
dnajb13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr4_-_16345227 | 1.07 |
ENSDART00000079521
|
kera
|
keratocan |
| chr19_-_15434813 | 1.06 |
ENSDART00000019843
|
ftr55
|
finTRIM family, member 55 |
| chr3_-_40955780 | 1.05 |
ENSDART00000130130
|
cyp3c3
|
cytochrome P450, family 3, subfamily c, polypeptide 3 |
| chr1_-_58562129 | 1.04 |
ENSDART00000159070
|
slc27a1b
|
solute carrier family 27 (fatty acid transporter), member 1b |
| chr17_-_42213822 | 1.03 |
ENSDART00000187904
ENSDART00000180029 |
nkx2.2a
|
NK2 homeobox 2a |
| chr12_-_31484677 | 1.01 |
ENSDART00000066578
|
tectb
|
tectorin beta |
| chr5_+_51079504 | 1.01 |
ENSDART00000097466
|
fam169aa
|
family with sequence similarity 169, member Aa |
| chr13_+_3954715 | 1.00 |
ENSDART00000182477
ENSDART00000192142 ENSDART00000190962 |
lrrc73
|
leucine rich repeat containing 73 |
| chr9_-_34882516 | 0.97 |
ENSDART00000011163
|
asmtl
|
acetylserotonin O-methyltransferase-like |
| chr7_-_58130703 | 0.97 |
ENSDART00000172082
|
ank2b
|
ankyrin 2b, neuronal |
| chr9_+_48219111 | 0.95 |
ENSDART00000111225
ENSDART00000145972 |
ccdc173
|
coiled-coil domain containing 173 |
| chr14_-_2318590 | 0.93 |
ENSDART00000192735
|
pcdh2ab8
|
protocadherin 2 alpha b 8 |
| chr2_-_5404466 | 0.92 |
ENSDART00000152907
|
si:ch1073-184j22.2
|
si:ch1073-184j22.2 |
| chr3_+_57779673 | 0.91 |
ENSDART00000101839
|
tmc6a
|
transmembrane channel-like 6a |
| chr2_+_33335911 | 0.89 |
ENSDART00000126135
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr20_+_22067337 | 0.86 |
ENSDART00000152636
|
clocka
|
clock circadian regulator a |
| chr25_-_17528994 | 0.86 |
ENSDART00000061712
|
si:dkey-44k1.5
|
si:dkey-44k1.5 |
| chr1_-_46632948 | 0.85 |
ENSDART00000148893
ENSDART00000053232 |
cdadc1
|
cytidine and dCMP deaminase domain containing 1 |
| chr19_-_18626952 | 0.84 |
ENSDART00000168004
ENSDART00000162034 ENSDART00000165486 ENSDART00000167971 |
rps18
|
ribosomal protein S18 |
| chr7_+_58699900 | 0.80 |
ENSDART00000144009
|
sdr16c5b
|
short chain dehydrogenase/reductase family 16C, member 5b |
| chr23_+_27779452 | 0.78 |
ENSDART00000134785
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr22_+_21317597 | 0.78 |
ENSDART00000132605
|
shc2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr1_+_37752171 | 0.78 |
ENSDART00000183247
ENSDART00000189756 ENSDART00000139448 |
GALNTL6
|
si:ch211-15e22.3 |
| chr14_+_24042760 | 0.77 |
ENSDART00000106096
|
drd1a
|
dopamine receptor D1a |
| chr2_+_50626476 | 0.76 |
ENSDART00000018150
|
neurod6b
|
neuronal differentiation 6b |
| chr13_-_14269626 | 0.75 |
ENSDART00000079176
|
gfra4a
|
GDNF family receptor alpha 4a |
| chr2_-_30770736 | 0.73 |
ENSDART00000131230
|
rgs20
|
regulator of G protein signaling 20 |
| chr2_-_4797512 | 0.72 |
ENSDART00000160765
|
tnk2b
|
tyrosine kinase, non-receptor, 2b |
| chr21_+_39365920 | 0.71 |
ENSDART00000140940
|
cluhb
|
clustered mitochondria (cluA/CLU1) homolog b |
| chr24_-_23839647 | 0.70 |
ENSDART00000125190
|
rrs1
|
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae) |
| chr17_+_15882533 | 0.70 |
ENSDART00000164124
|
gabrr2a
|
gamma-aminobutyric acid (GABA) A receptor, rho 2a |
| chr5_-_43071058 | 0.67 |
ENSDART00000165546
|
si:dkey-245n4.2
|
si:dkey-245n4.2 |
| chr6_-_35310224 | 0.66 |
ENSDART00000148997
|
nos1apa
|
nitric oxide synthase 1 (neuronal) adaptor protein a |
| chr16_+_50755133 | 0.66 |
ENSDART00000029283
|
IGLON5
|
zgc:110372 |
| chr18_-_12416019 | 0.65 |
ENSDART00000144799
|
si:ch211-1e14.1
|
si:ch211-1e14.1 |
| chr7_-_41964877 | 0.65 |
ENSDART00000092351
ENSDART00000193395 ENSDART00000187947 |
neto2b
|
neuropilin (NRP) and tolloid (TLL)-like 2b |
| chr1_+_1941031 | 0.61 |
ENSDART00000110331
|
PTGFRN
|
si:ch211-132g1.7 |
| chr17_-_13072334 | 0.60 |
ENSDART00000159598
|
CU469462.1
|
|
| chr7_+_18176162 | 0.60 |
ENSDART00000109171
|
rce1a
|
Ras converting CAAX endopeptidase 1a |
| chr7_+_48297842 | 0.59 |
ENSDART00000052123
|
slc25a44b
|
solute carrier family 25, member 44 b |
| chr16_-_46567136 | 0.59 |
ENSDART00000159180
|
si:dkey-152b24.7
|
si:dkey-152b24.7 |
| chr21_-_36396334 | 0.59 |
ENSDART00000183627
|
mrpl22
|
mitochondrial ribosomal protein L22 |
| chr8_+_7801060 | 0.58 |
ENSDART00000161618
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr25_+_19695652 | 0.57 |
ENSDART00000104340
|
mxg
|
myxovirus (influenza virus) resistance G |
| chr4_+_288633 | 0.52 |
ENSDART00000183304
|
FO834823.1
|
|
| chr12_+_23991276 | 0.52 |
ENSDART00000153136
|
psme4b
|
proteasome activator subunit 4b |
| chr2_+_30465102 | 0.50 |
ENSDART00000188404
|
neto1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr8_-_3346692 | 0.50 |
ENSDART00000057874
|
FUT9 (1 of many)
|
zgc:103510 |
| chr19_-_6083761 | 0.49 |
ENSDART00000151185
ENSDART00000143941 |
gsk3aa
|
glycogen synthase kinase 3 alpha a |
| chr21_-_30714665 | 0.49 |
ENSDART00000128011
|
tnfsf10l3
|
tumor necrosis factor (ligand) superfamily, member 10 like 3 |
| chr9_+_40874194 | 0.46 |
ENSDART00000141548
|
hibch
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr18_+_7264961 | 0.46 |
ENSDART00000188461
|
CABZ01015105.1
|
|
| chr23_-_33944597 | 0.41 |
ENSDART00000133223
|
si:dkey-190g6.2
|
si:dkey-190g6.2 |
| chr22_+_26853254 | 0.41 |
ENSDART00000182487
|
tmem186
|
transmembrane protein 186 |
| chr19_+_37135700 | 0.38 |
ENSDART00000103159
|
smim12
|
small integral membrane protein 12 |
| chr18_+_27511976 | 0.37 |
ENSDART00000132017
ENSDART00000140781 |
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr22_+_19366866 | 0.36 |
ENSDART00000137301
|
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr9_+_11034314 | 0.35 |
ENSDART00000032695
|
asic4a
|
acid-sensing (proton-gated) ion channel family member 4a |
| chr2_-_11662851 | 0.25 |
ENSDART00000145108
|
zgc:110130
|
zgc:110130 |
| chr5_+_37837245 | 0.25 |
ENSDART00000171617
|
epd
|
ependymin |
| chr18_+_5308392 | 0.24 |
ENSDART00000179072
|
DUT
|
deoxyuridine triphosphatase |
| chr3_+_15773991 | 0.21 |
ENSDART00000089923
|
znf652
|
zinc finger protein 652 |
| chr2_-_20666920 | 0.19 |
ENSDART00000143437
ENSDART00000114546 ENSDART00000136113 ENSDART00000179247 |
dusp12
|
dual specificity phosphatase 12 |
| chr12_-_3962372 | 0.18 |
ENSDART00000016791
|
eif3c
|
eukaryotic translation initiation factor 3, subunit C |
| chr20_+_34717403 | 0.17 |
ENSDART00000034252
|
pnocb
|
prepronociceptin b |
| chr12_+_1592146 | 0.15 |
ENSDART00000184575
ENSDART00000192902 |
SLC39A11
|
solute carrier family 39 member 11 |
| chr11_-_10850936 | 0.13 |
ENSDART00000091901
|
psmd14
|
proteasome 26S subunit, non-ATPase 14 |
| chr11_+_24851671 | 0.11 |
ENSDART00000167659
|
ipo9
|
importin 9 |
| chr17_+_2130018 | 0.07 |
ENSDART00000193675
ENSDART00000110529 |
bub1bb
|
BUB1 mitotic checkpoint serine/threonine kinase Bb |
| chr1_-_45889820 | 0.05 |
ENSDART00000144735
|
pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr17_+_7595356 | 0.04 |
ENSDART00000130625
|
si:dkeyp-110a12.4
|
si:dkeyp-110a12.4 |
| chr2_-_30784198 | 0.04 |
ENSDART00000182523
ENSDART00000147355 |
rgs20
|
regulator of G protein signaling 20 |
Network of associatons between targets according to the STRING database.
First level regulatory network of six4a+six4b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:1903373 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.8 | 2.3 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.6 | 2.5 | GO:0072149 | visceral serous pericardium development(GO:0061032) glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.5 | 2.6 | GO:0031652 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.5 | 2.3 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.4 | 2.6 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.4 | 1.6 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.3 | 2.3 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.3 | 5.1 | GO:0014823 | response to activity(GO:0014823) |
| 0.2 | 1.9 | GO:0060005 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.2 | 1.6 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.2 | 0.9 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.2 | 2.2 | GO:0015813 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.2 | 1.8 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 0.6 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.2 | 1.0 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.2 | 2.0 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.2 | 1.7 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.2 | 1.4 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 2.6 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.7 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.1 | 0.8 | GO:0042311 | vasodilation(GO:0042311) |
| 0.1 | 8.6 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 1.7 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.1 | 0.6 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 1.6 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.8 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 0.5 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 0.5 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 2.0 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 0.7 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.6 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 1.1 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 1.7 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.5 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 1.9 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 1.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 1.4 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 1.5 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 1.5 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.4 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.9 | GO:0009266 | response to temperature stimulus(GO:0009266) |
| 0.0 | 1.3 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 2.0 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 1.0 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.5 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 4.0 | GO:0006520 | cellular amino acid metabolic process(GO:0006520) |
| 0.0 | 1.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.6 | GO:0042255 | ribosome assembly(GO:0042255) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.2 | 0.9 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.2 | 2.0 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 1.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.6 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 1.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 3.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.8 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.7 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 1.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 2.1 | GO:0015935 | small ribosomal subunit(GO:0015935) |
| 0.0 | 0.7 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 2.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.7 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 2.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.0 | GO:0034703 | cation channel complex(GO:0034703) |
| 0.0 | 6.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.5 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.6 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 3.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.4 | 2.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.4 | 4.0 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.3 | 1.6 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.3 | 0.8 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.2 | 5.1 | GO:0005344 | oxygen transporter activity(GO:0005344) oxygen binding(GO:0019825) |
| 0.2 | 8.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.2 | 2.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 1.5 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 2.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.2 | 1.0 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.2 | 1.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 2.6 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 0.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.9 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.7 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 2.0 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 2.0 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 2.8 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 21.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 0.9 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 0.9 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 1.6 | GO:0016411 | acylglycerol O-acyltransferase activity(GO:0016411) |
| 0.1 | 2.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.9 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.1 | 0.5 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 1.0 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.4 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 2.1 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.8 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 2.7 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 1.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.4 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 1.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.5 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 2.1 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 3.8 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.5 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.6 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 1.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.8 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 1.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.6 | PID SHP2 PATHWAY | SHP2 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 1.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 0.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.8 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 2.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.6 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 0.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |