Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
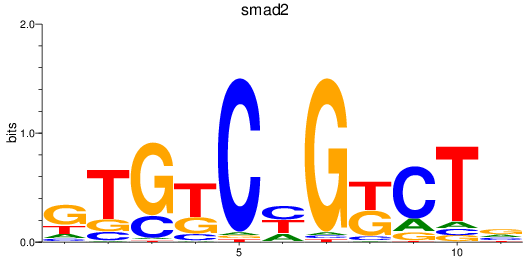
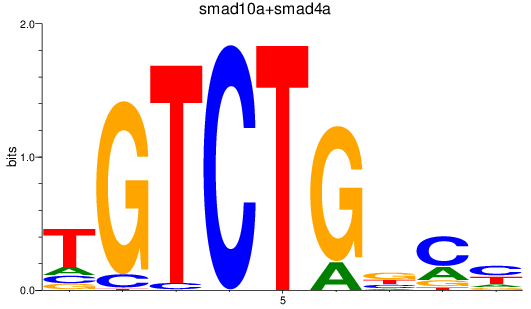
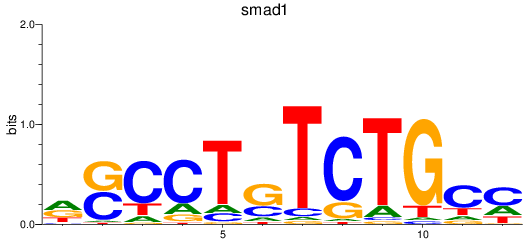
Results for smad2_smad10a+smad4a_smad1
Z-value: 0.97
Transcription factors associated with smad2_smad10a+smad4a_smad1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
smad2
|
ENSDARG00000006389 | SMAD family member 2 |
|
smad10a
|
ENSDARG00000045094 | si_dkey-222b8.1 |
|
smad10a
|
ENSDARG00000070428 | si_dkey-222b8.1 |
|
smad4a
|
ENSDARG00000075226 | SMAD family member 4a |
|
smad1
|
ENSDARG00000027199 | SMAD family member 1 |
|
smad1
|
ENSDARG00000112617 | SMAD family member 1 |
|
smad1
|
ENSDARG00000115674 | SMAD family member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| smad4a | dr11_v1_chr10_+_575929_575929 | 0.63 | 7.7e-12 | Click! |
| si:dkey-239n17.4 | dr11_v1_chr16_-_28876479_28876479 | 0.45 | 6.0e-06 | Click! |
| smad2 | dr11_v1_chr10_-_14929630_14929679 | -0.14 | 1.8e-01 | Click! |
| smad1 | dr11_v1_chr1_-_35916247_35916247 | 0.12 | 2.3e-01 | Click! |
| si:dkey-222b8.1 | dr11_v1_chr19_+_23932259_23932259 | -0.06 | 5.7e-01 | Click! |
Activity profile of smad2_smad10a+smad4a_smad1 motif
Sorted Z-values of smad2_smad10a+smad4a_smad1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_3496738 | 8.30 |
ENSDART00000186849
|
CABZ01040998.1
|
|
| chr3_+_26813058 | 7.58 |
ENSDART00000055537
|
socs1a
|
suppressor of cytokine signaling 1a |
| chr9_-_8670158 | 7.39 |
ENSDART00000077296
|
col4a1
|
collagen, type IV, alpha 1 |
| chr22_-_557965 | 6.88 |
ENSDART00000001201
|
bysl
|
bystin-like |
| chr11_+_45255774 | 6.31 |
ENSDART00000172838
|
aspscr1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr17_+_33433576 | 5.92 |
ENSDART00000077581
|
snap23.2
|
synaptosomal-associated protein 23.2 |
| chr5_-_3960161 | 5.71 |
ENSDART00000111453
|
myo19
|
myosin XIX |
| chr4_-_16412084 | 5.58 |
ENSDART00000188460
|
dcn
|
decorin |
| chr6_-_31827597 | 5.52 |
ENSDART00000159400
|
ror1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr4_-_21466825 | 5.42 |
ENSDART00000066897
|
pawr
|
PRKC, apoptosis, WT1, regulator |
| chr13_-_479129 | 5.36 |
ENSDART00000159803
ENSDART00000082127 |
heatr5b
|
HEAT repeat containing 5B |
| chr15_+_28303161 | 5.18 |
ENSDART00000087926
|
myo1cb
|
myosin Ic, paralog b |
| chr4_-_4932619 | 5.03 |
ENSDART00000103293
|
ndufa5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr5_+_64900223 | 5.03 |
ENSDART00000191677
|
ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr12_+_15165736 | 4.76 |
ENSDART00000180398
|
adprh
|
ADP-ribosylarginine hydrolase |
| chr20_-_15922986 | 4.49 |
ENSDART00000189421
|
fam20b
|
family with sequence similarity 20, member B (H. sapiens) |
| chr10_+_25272355 | 4.41 |
ENSDART00000140186
|
rab38b
|
RAB38b, member of RAS oncogene family |
| chr19_-_9648542 | 3.74 |
ENSDART00000172628
|
clcn1a
|
chloride channel, voltage-sensitive 1a |
| chr25_-_774350 | 3.57 |
ENSDART00000166321
ENSDART00000160386 |
IRAK4
|
interleukin 1 receptor associated kinase 4 |
| chr18_-_5542351 | 3.57 |
ENSDART00000129459
|
spata5l1
|
spermatogenesis associated 5-like 1 |
| chr13_-_1130096 | 3.56 |
ENSDART00000010261
|
pno1
|
partner of NOB1 homolog |
| chr19_-_5380770 | 3.50 |
ENSDART00000000221
|
krt91
|
keratin 91 |
| chr10_-_10018120 | 3.48 |
ENSDART00000132375
|
strbp
|
spermatid perinuclear RNA binding protein |
| chr4_-_25257451 | 3.46 |
ENSDART00000142819
|
tmem110l
|
transmembrane protein 110, like |
| chr20_+_54356540 | 3.39 |
ENSDART00000143591
|
znf410
|
zinc finger protein 410 |
| chr22_-_506522 | 3.30 |
ENSDART00000106645
ENSDART00000067637 |
dstyk
|
dual serine/threonine and tyrosine protein kinase |
| chr22_+_35068046 | 3.26 |
ENSDART00000161660
ENSDART00000169573 |
si:ch73-173h19.3
|
si:ch73-173h19.3 |
| chr23_-_45405968 | 3.23 |
ENSDART00000149462
|
zgc:101853
|
zgc:101853 |
| chr12_-_4070058 | 3.23 |
ENSDART00000042200
|
aldoab
|
aldolase a, fructose-bisphosphate, b |
| chr11_+_77526 | 3.17 |
ENSDART00000193521
|
CABZ01072242.1
|
|
| chr19_+_19988869 | 3.17 |
ENSDART00000151024
|
osbpl3a
|
oxysterol binding protein-like 3a |
| chr2_-_42628028 | 3.14 |
ENSDART00000179866
|
myo10
|
myosin X |
| chr18_-_48755037 | 3.11 |
ENSDART00000109673
|
tirap
|
toll-interleukin 1 receptor (TIR) domain containing adaptor protein |
| chr19_-_43819582 | 2.96 |
ENSDART00000160879
ENSDART00000075902 |
klhl43
|
kelch-like family member 43 |
| chr18_+_5549672 | 2.95 |
ENSDART00000184970
|
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr4_-_797831 | 2.87 |
ENSDART00000158970
ENSDART00000170012 |
mapre3b
|
microtubule-associated protein, RP/EB family, member 3b |
| chr7_+_22767678 | 2.84 |
ENSDART00000137203
|
ponzr6
|
plac8 onzin related protein 6 |
| chr14_-_237130 | 2.83 |
ENSDART00000164988
|
bod1l1
|
biorientation of chromosomes in cell division 1-like 1 |
| chr2_-_42705284 | 2.71 |
ENSDART00000187160
|
myo10
|
myosin X |
| chr9_-_180334 | 2.70 |
ENSDART00000180339
|
zgc:158619
|
zgc:158619 |
| chr25_-_31396479 | 2.69 |
ENSDART00000156828
|
prr33
|
proline rich 33 |
| chr12_+_30653047 | 2.69 |
ENSDART00000148562
|
thbs2b
|
thrombospondin 2b |
| chr3_+_32526799 | 2.66 |
ENSDART00000185755
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr11_+_42585138 | 2.60 |
ENSDART00000019008
|
asb14a
|
ankyrin repeat and SOCS box containing 14a |
| chr20_-_15922210 | 2.56 |
ENSDART00000152412
ENSDART00000152354 ENSDART00000152828 ENSDART00000013453 ENSDART00000152357 |
fam20b
|
family with sequence similarity 20, member B (H. sapiens) |
| chr7_+_56615554 | 2.54 |
ENSDART00000098430
|
dpep1
|
dipeptidase 1 |
| chr3_+_32526263 | 2.41 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr21_-_45073 | 2.34 |
ENSDART00000185997
|
bhmt
|
betaine-homocysteine methyltransferase |
| chr20_+_23625387 | 2.30 |
ENSDART00000147945
ENSDART00000150497 |
palld
|
palladin, cytoskeletal associated protein |
| chr3_+_41714966 | 2.22 |
ENSDART00000155440
|
eif3ba
|
eukaryotic translation initiation factor 3, subunit Ba |
| chr14_-_25949951 | 2.21 |
ENSDART00000141304
|
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr3_-_21242460 | 2.21 |
ENSDART00000007293
|
tcap
|
titin-cap (telethonin) |
| chr5_-_72125551 | 2.17 |
ENSDART00000149412
|
smyd1a
|
SET and MYND domain containing 1a |
| chr22_+_635813 | 2.17 |
ENSDART00000179067
|
CU856139.1
|
|
| chr14_+_51098036 | 2.16 |
ENSDART00000184118
|
CABZ01078594.1
|
|
| chr25_+_32496877 | 2.15 |
ENSDART00000132698
|
ctdspl2a
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2a |
| chr24_-_28243186 | 2.15 |
ENSDART00000105691
|
cox4i1l
|
cytochrome c oxidase subunit IV isoform 1, like |
| chr18_+_50961953 | 2.13 |
ENSDART00000158768
|
ppfia1
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 |
| chr1_+_59073203 | 2.09 |
ENSDART00000149937
ENSDART00000162201 |
MFAP4 (1 of many)
zgc:173915
|
si:zfos-2330d3.3 zgc:173915 |
| chr19_+_41551335 | 2.08 |
ENSDART00000169193
|
si:ch211-57n23.4
|
si:ch211-57n23.4 |
| chr24_+_38306010 | 2.05 |
ENSDART00000143184
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr16_+_40301056 | 2.01 |
ENSDART00000058578
|
rspo3
|
R-spondin 3 |
| chr9_-_35875051 | 1.96 |
ENSDART00000013432
|
dnajc28
|
DnaJ (Hsp40) homolog, subfamily C, member 28 |
| chr8_+_32719930 | 1.95 |
ENSDART00000145362
|
hmcn2
|
hemicentin 2 |
| chr4_-_1015896 | 1.93 |
ENSDART00000170292
|
FAM180A
|
family with sequence similarity 180 member A |
| chr4_-_17785120 | 1.90 |
ENSDART00000024775
|
mybpc1
|
myosin binding protein C, slow type |
| chr4_+_77993103 | 1.86 |
ENSDART00000174392
|
terfa
|
telomeric repeat binding factor a |
| chr2_-_37098785 | 1.81 |
ENSDART00000003670
|
zgc:101744
|
zgc:101744 |
| chr5_-_34997630 | 1.77 |
ENSDART00000170684
|
btf3
|
basic transcription factor 3 |
| chr13_+_51981764 | 1.77 |
ENSDART00000160698
|
klhdc3
|
kelch domain containing 3 |
| chr23_-_32156278 | 1.73 |
ENSDART00000157479
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr5_+_27898226 | 1.69 |
ENSDART00000098604
ENSDART00000180251 |
adam28
|
ADAM metallopeptidase domain 28 |
| chr3_+_30257582 | 1.68 |
ENSDART00000159497
ENSDART00000103457 ENSDART00000121883 |
mybpc2a
|
myosin binding protein C, fast type a |
| chr23_+_36460239 | 1.68 |
ENSDART00000172441
|
lima1a
|
LIM domain and actin binding 1a |
| chr17_-_122680 | 1.65 |
ENSDART00000066430
|
actc1b
|
actin, alpha, cardiac muscle 1b |
| chr14_-_6987649 | 1.64 |
ENSDART00000060990
|
eif4ebp3l
|
eukaryotic translation initiation factor 4E binding protein 3, like |
| chr19_+_48111285 | 1.63 |
ENSDART00000169420
|
nme2b.2
|
NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 2 |
| chr20_+_18163355 | 1.62 |
ENSDART00000011287
|
aqp4
|
aquaporin 4 |
| chr8_-_14126646 | 1.62 |
ENSDART00000027225
|
bgna
|
biglycan a |
| chr6_-_15653494 | 1.60 |
ENSDART00000038133
|
trim63a
|
tripartite motif containing 63a |
| chr10_+_35526528 | 1.58 |
ENSDART00000184110
|
phldb2a
|
pleckstrin homology-like domain, family B, member 2a |
| chr19_+_4061699 | 1.56 |
ENSDART00000158309
ENSDART00000166512 |
btr25
btr26
|
bloodthirsty-related gene family, member 25 bloodthirsty-related gene family, member 26 |
| chr2_+_58739111 | 1.56 |
ENSDART00000097690
|
CABZ01083448.1
|
|
| chr9_+_23900703 | 1.55 |
ENSDART00000127859
|
trim63b
|
tripartite motif containing 63b |
| chr23_+_25995727 | 1.54 |
ENSDART00000137123
|
wisp2
|
WNT1 inducible signaling pathway protein 2 |
| chr16_-_11961325 | 1.52 |
ENSDART00000143845
|
si:ch73-296e2.3
|
si:ch73-296e2.3 |
| chr24_-_24281792 | 1.51 |
ENSDART00000146482
ENSDART00000018420 |
pdha1b
|
pyruvate dehydrogenase E1 alpha 1 subunit b |
| chr7_-_18508815 | 1.49 |
ENSDART00000173539
|
rgs12a
|
regulator of G protein signaling 12a |
| chr3_-_31019715 | 1.46 |
ENSDART00000156687
ENSDART00000156127 ENSDART00000153945 |
si:dkeyp-71f10.5
|
si:dkeyp-71f10.5 |
| chr6_-_49526510 | 1.45 |
ENSDART00000128025
|
rps26l
|
ribosomal protein S26, like |
| chr18_-_44908479 | 1.44 |
ENSDART00000169636
|
si:ch211-71n6.4
|
si:ch211-71n6.4 |
| chr9_-_34191627 | 1.44 |
ENSDART00000142664
|
dcaf6
|
ddb1 and cul4 associated factor 6 |
| chr20_+_7084154 | 1.40 |
ENSDART00000136448
|
ftr85
|
finTRIM family, member 85 |
| chr14_-_736575 | 1.39 |
ENSDART00000168611
|
tlr1
|
toll-like receptor 1 |
| chr8_-_40070114 | 1.39 |
ENSDART00000036280
|
ggt1a
|
gamma-glutamyltransferase 1a |
| chr1_+_1805294 | 1.38 |
ENSDART00000103850
|
atp1a1a.3
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 3 |
| chr17_-_15546862 | 1.38 |
ENSDART00000091021
|
col10a1a
|
collagen, type X, alpha 1a |
| chr4_+_55758103 | 1.37 |
ENSDART00000185964
|
CT583728.23
|
|
| chr19_+_43119698 | 1.37 |
ENSDART00000167847
ENSDART00000186962 ENSDART00000187305 |
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr19_-_81851 | 1.37 |
ENSDART00000172319
|
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr2_-_2642476 | 1.35 |
ENSDART00000124032
|
serbp1b
|
SERPINE1 mRNA binding protein 1b |
| chr20_-_2355357 | 1.32 |
ENSDART00000085281
|
EPB41L2
|
si:ch73-18b11.1 |
| chr12_-_17712393 | 1.32 |
ENSDART00000143534
ENSDART00000010144 |
pvalb2
|
parvalbumin 2 |
| chr7_-_69857692 | 1.32 |
ENSDART00000124764
|
myoz2a
|
myozenin 2a |
| chr9_+_310331 | 1.30 |
ENSDART00000172446
ENSDART00000187731 ENSDART00000193970 |
stac3
|
SH3 and cysteine rich domain 3 |
| chr21_+_5993188 | 1.29 |
ENSDART00000048399
|
slc4a4b
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4b |
| chr13_+_22480496 | 1.27 |
ENSDART00000136863
ENSDART00000131870 ENSDART00000078720 ENSDART00000078740 ENSDART00000139218 |
ldb3a
|
LIM domain binding 3a |
| chr11_-_13152524 | 1.27 |
ENSDART00000181440
ENSDART00000191997 |
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr21_+_30351256 | 1.25 |
ENSDART00000078341
|
foxi3a
|
forkhead box I3a |
| chr23_+_19790962 | 1.25 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr6_-_29195642 | 1.24 |
ENSDART00000078625
|
dpt
|
dermatopontin |
| chr23_-_46040618 | 1.24 |
ENSDART00000161415
|
CABZ01080918.1
|
|
| chr1_-_681116 | 1.23 |
ENSDART00000165894
|
adamts1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr3_-_29928695 | 1.23 |
ENSDART00000151275
ENSDART00000151083 |
grna
|
granulin a |
| chr22_+_3303671 | 1.23 |
ENSDART00000075049
|
gipc3
|
GIPC PDZ domain containing family, member 3 |
| chr10_-_35149513 | 1.21 |
ENSDART00000063434
ENSDART00000131291 |
ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr9_+_44304980 | 1.21 |
ENSDART00000147990
|
ssfa2
|
sperm specific antigen 2 |
| chr17_+_24843401 | 1.20 |
ENSDART00000110179
|
cx34.4
|
connexin 34.4 |
| chr21_+_33454147 | 1.20 |
ENSDART00000053208
|
rps14
|
ribosomal protein S14 |
| chr20_+_50061890 | 1.20 |
ENSDART00000137725
|
cpsf2
|
cleavage and polyadenylation specific factor 2 |
| chr24_-_4768955 | 1.19 |
ENSDART00000066834
|
agtr1b
|
angiotensin II receptor, type 1b |
| chr3_+_13879446 | 1.19 |
ENSDART00000164767
|
farsa
|
phenylalanyl-tRNA synthetase, alpha subunit |
| chr16_-_26132122 | 1.16 |
ENSDART00000157787
|
lipeb
|
lipase, hormone-sensitive b |
| chr15_-_28082310 | 1.16 |
ENSDART00000152620
|
dhrs13a.3
|
dehydrogenase/reductase (SDR family) member 13a, duplicate 3 |
| chr21_+_10866421 | 1.15 |
ENSDART00000137858
|
alpk2
|
alpha-kinase 2 |
| chr5_+_34997763 | 1.15 |
ENSDART00000043341
|
foxd1
|
forkhead box D1 |
| chr19_-_82504 | 1.15 |
ENSDART00000027864
ENSDART00000160560 |
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr20_+_54356272 | 1.15 |
ENSDART00000145735
|
znf410
|
zinc finger protein 410 |
| chr25_-_6261693 | 1.14 |
ENSDART00000135808
|
ireb2
|
iron-responsive element binding protein 2 |
| chr19_-_38611814 | 1.12 |
ENSDART00000151958
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr22_+_18886209 | 1.11 |
ENSDART00000144402
|
fstl3
|
follistatin-like 3 (secreted glycoprotein) |
| chr1_+_36552 | 1.10 |
ENSDART00000169685
|
hikeshi
|
Hikeshi, heat shock protein nuclear import factor |
| chr9_-_23922011 | 1.10 |
ENSDART00000145734
|
col6a3
|
collagen, type VI, alpha 3 |
| chr3_-_48980319 | 1.08 |
ENSDART00000165319
|
ftr42
|
finTRIM family, member 42 |
| chr25_+_245018 | 1.08 |
ENSDART00000155344
|
zgc:92481
|
zgc:92481 |
| chr3_+_5331428 | 1.08 |
ENSDART00000156561
|
si:ch73-106l15.4
|
si:ch73-106l15.4 |
| chr5_+_8964926 | 1.07 |
ENSDART00000091397
ENSDART00000164535 |
tnksb
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase b |
| chr14_-_12307522 | 1.06 |
ENSDART00000163900
|
myot
|
myotilin |
| chr21_-_11632403 | 1.06 |
ENSDART00000171708
ENSDART00000138619 ENSDART00000136308 ENSDART00000144770 |
cast
|
calpastatin |
| chr24_+_6353394 | 1.05 |
ENSDART00000165118
|
CR352329.1
|
|
| chr23_-_17470146 | 1.04 |
ENSDART00000002398
|
trim101
|
tripartite motif containing 101 |
| chr5_-_1962500 | 1.04 |
ENSDART00000150163
|
rplp0
|
ribosomal protein, large, P0 |
| chr7_+_9290929 | 1.04 |
ENSDART00000128530
|
snrpa1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr4_+_41602 | 1.03 |
ENSDART00000159640
|
phtf2
|
putative homeodomain transcription factor 2 |
| chr19_-_3574060 | 1.03 |
ENSDART00000105120
|
tmem170b
|
transmembrane protein 170B |
| chr13_-_36418921 | 1.02 |
ENSDART00000135804
|
dcaf5
|
ddb1 and cul4 associated factor 5 |
| chr19_+_4062101 | 1.01 |
ENSDART00000166773
|
btr25
|
bloodthirsty-related gene family, member 25 |
| chr18_+_30847237 | 1.01 |
ENSDART00000012374
|
foxf1
|
forkhead box F1 |
| chr15_-_47468085 | 1.01 |
ENSDART00000164438
|
inppl1a
|
inositol polyphosphate phosphatase-like 1a |
| chr12_+_33894396 | 1.00 |
ENSDART00000130853
ENSDART00000152988 |
mfsd13a
|
major facilitator superfamily domain containing 13A |
| chr20_+_46544052 | 0.99 |
ENSDART00000098968
|
si:ch211-153j24.3
|
si:ch211-153j24.3 |
| chr20_-_31427390 | 0.99 |
ENSDART00000007735
|
ust
|
uronyl-2-sulfotransferase |
| chr6_-_60104628 | 0.99 |
ENSDART00000057463
ENSDART00000169188 |
pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr11_-_141592 | 0.99 |
ENSDART00000092787
|
cdk4
|
cyclin-dependent kinase 4 |
| chr11_+_141504 | 0.99 |
ENSDART00000086166
|
NCKAP1L
|
zgc:172352 |
| chr25_+_37340722 | 0.98 |
ENSDART00000137025
|
pamr1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr16_+_54588930 | 0.98 |
ENSDART00000159174
|
dennd4b
|
DENN/MADD domain containing 4B |
| chr25_-_3892686 | 0.98 |
ENSDART00000043172
|
tmem258
|
transmembrane protein 258 |
| chr5_-_33286820 | 0.98 |
ENSDART00000184426
|
rpl7a
|
ribosomal protein L7a |
| chr20_+_34400715 | 0.97 |
ENSDART00000061632
|
fam129aa
|
family with sequence similarity 129, member Aa |
| chr2_+_13069168 | 0.97 |
ENSDART00000192832
|
prkag2b
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit b |
| chr22_+_661505 | 0.95 |
ENSDART00000149460
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr6_-_24103666 | 0.95 |
ENSDART00000164915
|
scinla
|
scinderin like a |
| chr12_-_49165751 | 0.95 |
ENSDART00000148491
ENSDART00000112479 |
acadsb
|
acyl-CoA dehydrogenase short/branched chain |
| chr22_-_18779232 | 0.95 |
ENSDART00000186726
|
atp5f1d
|
ATP synthase F1 subunit delta |
| chr24_+_39990695 | 0.93 |
ENSDART00000040281
|
BX323854.1
|
|
| chr5_+_483965 | 0.93 |
ENSDART00000150007
|
tek
|
TEK tyrosine kinase, endothelial |
| chr14_+_11458044 | 0.93 |
ENSDART00000186425
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr12_-_5505205 | 0.93 |
ENSDART00000092319
|
abi3b
|
ABI family, member 3b |
| chr3_-_1388936 | 0.93 |
ENSDART00000171278
|
ddx47
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 |
| chr22_+_26400519 | 0.92 |
ENSDART00000159839
ENSDART00000144585 |
capn8
|
calpain 8 |
| chr10_+_29259882 | 0.92 |
ENSDART00000180606
|
sytl2a
|
synaptotagmin-like 2a |
| chr13_+_22480857 | 0.91 |
ENSDART00000078721
ENSDART00000044719 ENSDART00000130957 ENSDART00000078757 ENSDART00000130424 ENSDART00000078747 |
ldb3a
|
LIM domain binding 3a |
| chr4_+_77933084 | 0.91 |
ENSDART00000148728
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr23_-_24263474 | 0.91 |
ENSDART00000160312
|
hspb7
|
heat shock protein family, member 7 (cardiovascular) |
| chr19_-_5369486 | 0.90 |
ENSDART00000105004
|
krt17
|
keratin 17 |
| chr5_-_58780160 | 0.90 |
ENSDART00000166955
|
arhgef12b
|
Rho guanine nucleotide exchange factor (GEF) 12b |
| chr23_+_43200911 | 0.90 |
ENSDART00000164262
|
si:dkey-65j6.2
|
si:dkey-65j6.2 |
| chr17_-_52091999 | 0.89 |
ENSDART00000019766
|
tgfb3
|
transforming growth factor, beta 3 |
| chr6_+_103361 | 0.89 |
ENSDART00000151899
|
ldlrb
|
low density lipoprotein receptor b |
| chr13_-_280652 | 0.88 |
ENSDART00000193627
|
slc30a6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr8_-_28304534 | 0.88 |
ENSDART00000078545
|
lmod1a
|
leiomodin 1a (smooth muscle) |
| chr9_+_307863 | 0.87 |
ENSDART00000163474
|
stac3
|
SH3 and cysteine rich domain 3 |
| chr3_-_1434135 | 0.87 |
ENSDART00000149622
|
mgp
|
matrix Gla protein |
| chr11_+_42556395 | 0.87 |
ENSDART00000039206
|
rps23
|
ribosomal protein S23 |
| chr14_+_2243 | 0.87 |
ENSDART00000191193
|
CYTL1
|
cytokine like 1 |
| chr7_-_51433903 | 0.87 |
ENSDART00000083189
|
arhgap36
|
Rho GTPase activating protein 36 |
| chr12_-_628024 | 0.87 |
ENSDART00000158563
ENSDART00000152612 |
wu:fj29h11
si:ch73-301j1.1
|
wu:fj29h11 si:ch73-301j1.1 |
| chr19_+_47394270 | 0.86 |
ENSDART00000171281
|
psmb2
|
proteasome subunit beta 2 |
| chr22_+_38824012 | 0.85 |
ENSDART00000144318
ENSDART00000003736 |
anos1b
|
anosmin 1b |
| chr11_-_44999858 | 0.85 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr2_-_59265521 | 0.85 |
ENSDART00000146341
ENSDART00000097799 |
ftr33
|
finTRIM family, member 33 |
| chr11_-_44647286 | 0.84 |
ENSDART00000169329
ENSDART00000158939 |
tomm20b
|
translocase of outer mitochondrial membrane 20b |
| chr22_+_661711 | 0.84 |
ENSDART00000113795
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr11_+_40032790 | 0.84 |
ENSDART00000158809
|
si:dkey-264d12.1
|
si:dkey-264d12.1 |
| chr14_-_52583021 | 0.84 |
ENSDART00000166924
|
CU469526.1
|
|
| chr5_+_18047111 | 0.84 |
ENSDART00000132164
|
hira
|
histone cell cycle regulator a |
| chr23_+_43201142 | 0.84 |
ENSDART00000158832
ENSDART00000190315 |
si:dkey-65j6.2
|
si:dkey-65j6.2 |
| chr8_+_554531 | 0.84 |
ENSDART00000193623
|
FO704758.2
|
|
| chr1_-_633356 | 0.84 |
ENSDART00000171019
|
appa
|
amyloid beta (A4) precursor protein a |
Network of associatons between targets according to the STRING database.
First level regulatory network of smad2_smad10a+smad4a_smad1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 1.3 | 7.6 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 1.3 | 5.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.7 | 2.9 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.6 | 2.2 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.5 | 2.0 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.5 | 1.9 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.4 | 2.2 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.4 | 1.6 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.4 | 1.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.4 | 1.2 | GO:0071831 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.4 | 3.3 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.4 | 2.8 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.3 | 6.2 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.3 | 2.9 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.3 | 1.3 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.3 | 16.7 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.3 | 1.3 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.3 | 2.0 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.3 | 1.1 | GO:1904355 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.3 | 0.8 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.3 | 6.9 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.3 | 0.3 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.2 | 2.2 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.2 | 0.7 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.2 | 0.7 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.2 | 0.7 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.2 | 1.6 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.2 | 1.3 | GO:0035989 | tendon development(GO:0035989) |
| 0.2 | 0.6 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.2 | 1.7 | GO:0031179 | peptide modification(GO:0031179) |
| 0.2 | 6.3 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.2 | 5.6 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.2 | 0.8 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.2 | 3.1 | GO:0003313 | heart rudiment development(GO:0003313) |
| 0.2 | 2.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 3.9 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.2 | 1.7 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.2 | 1.0 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.2 | 1.3 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 0.7 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.2 | 0.7 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.2 | 1.6 | GO:0006833 | water transport(GO:0006833) |
| 0.2 | 1.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 | 0.5 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.2 | 0.8 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.2 | 1.2 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.2 | 1.1 | GO:0051705 | multi-organism behavior(GO:0051705) |
| 0.2 | 0.5 | GO:1902893 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.2 | 2.3 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.2 | 0.5 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.2 | 0.8 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.2 | 0.5 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.2 | 1.2 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.2 | 0.6 | GO:0015840 | urea transport(GO:0015840) |
| 0.2 | 0.5 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.2 | 1.1 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 0.9 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.1 | 2.1 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.1 | 0.1 | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) |
| 0.1 | 1.0 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 0.4 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.1 | 1.4 | GO:0050729 | positive regulation of inflammatory response(GO:0050729) |
| 0.1 | 2.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.6 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 1.0 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.5 | GO:0072512 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.1 | 0.7 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 1.1 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 0.8 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.6 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.3 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.1 | 0.9 | GO:0001765 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 2.0 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 1.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 0.7 | GO:0097350 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 2.6 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.7 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 1.6 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 0.4 | GO:0061033 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.1 | 2.4 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.1 | 0.5 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 6.2 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.1 | 0.4 | GO:0033153 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.1 | 0.1 | GO:0048338 | axial mesoderm structural organization(GO:0048331) mesoderm structural organization(GO:0048338) |
| 0.1 | 0.4 | GO:0015865 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.1 | 0.3 | GO:0010658 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.1 | 0.5 | GO:0035776 | pronephric proximal tubule development(GO:0035776) |
| 0.1 | 0.4 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.1 | 5.4 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.1 | 0.3 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.1 | 0.2 | GO:0045064 | T-helper 2 cell differentiation(GO:0045064) |
| 0.1 | 0.7 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.1 | 1.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 5.4 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 0.3 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 0.6 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.1 | 0.2 | GO:0071314 | response to cocaine(GO:0042220) cellular response to alkaloid(GO:0071312) cellular response to cocaine(GO:0071314) |
| 0.1 | 0.5 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.1 | 1.5 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.1 | 1.9 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.8 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.6 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.1 | 1.5 | GO:0070672 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.1 | 0.6 | GO:0050779 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.1 | 0.7 | GO:2000377 | regulation of reactive oxygen species metabolic process(GO:2000377) |
| 0.1 | 0.4 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 0.2 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.8 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 | 0.2 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 0.8 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.1 | 0.3 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.1 | 0.3 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.1 | 0.8 | GO:0032925 | regulation of activin receptor signaling pathway(GO:0032925) regulation of nodal signaling pathway(GO:1900107) |
| 0.1 | 1.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.6 | GO:0010888 | negative regulation of cholesterol storage(GO:0010887) negative regulation of lipid storage(GO:0010888) |
| 0.1 | 0.8 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 0.3 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 0.4 | GO:0033986 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.1 | 0.2 | GO:0033632 | cell-cell adhesion mediated by integrin(GO:0033631) regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.1 | 2.9 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.1 | 0.5 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 2.0 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.2 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.1 | 0.2 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.1 | 0.6 | GO:0086002 | cardiac muscle cell action potential involved in contraction(GO:0086002) |
| 0.1 | 0.7 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.4 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 0.2 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 2.1 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.1 | 0.4 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 0.6 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 0.2 | GO:0032060 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.1 | 0.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 1.1 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 0.2 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 2.8 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.1 | 0.8 | GO:0070654 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.1 | 0.5 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 2.1 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.3 | GO:0039689 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.7 | GO:0060021 | palate development(GO:0060021) |
| 0.1 | 0.5 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 0.6 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 0.2 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.1 | 1.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.2 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.0 | 0.2 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.7 | GO:0042509 | tyrosine phosphorylation of STAT protein(GO:0007260) regulation of tyrosine phosphorylation of STAT protein(GO:0042509) |
| 0.0 | 0.2 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.0 | 0.3 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.7 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 1.5 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 1.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.7 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.4 | GO:0033273 | response to vitamin(GO:0033273) |
| 0.0 | 0.1 | GO:0044241 | intestinal cholesterol absorption(GO:0030299) lipid digestion(GO:0044241) |
| 0.0 | 0.3 | GO:0048662 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.0 | 2.2 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 5.9 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.3 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridine biosynthesis.(GO:0002926) |
| 0.0 | 0.3 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.2 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 0.9 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 1.2 | GO:0019229 | regulation of vasoconstriction(GO:0019229) |
| 0.0 | 0.2 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.3 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.4 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.2 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.5 | GO:2000142 | regulation of transcription initiation from RNA polymerase II promoter(GO:0060260) regulation of DNA-templated transcription, initiation(GO:2000142) |
| 0.0 | 0.9 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.1 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.5 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.2 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 0.3 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 1.0 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.6 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 5.8 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.2 | GO:1901073 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.0 | 0.4 | GO:0043536 | positive regulation of blood vessel endothelial cell migration(GO:0043536) positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.4 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 2.4 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.8 | GO:0043491 | protein kinase B signaling(GO:0043491) |
| 0.0 | 0.6 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.5 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.3 | GO:1901099 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.2 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.3 | GO:0046620 | regulation of organ growth(GO:0046620) |
| 0.0 | 0.2 | GO:0016127 | steroid catabolic process(GO:0006706) cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.4 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.3 | GO:0097531 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 2.9 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.8 | GO:0045104 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.2 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.7 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.2 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.0 | 2.3 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.4 | GO:0098754 | detoxification(GO:0098754) |
| 0.0 | 0.0 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 1.5 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.1 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.3 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.0 | 0.2 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.0 | 0.4 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.7 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 1.2 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.7 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.3 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 1.4 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 0.1 | GO:0060323 | head morphogenesis(GO:0060323) face morphogenesis(GO:0060325) |
| 0.0 | 1.4 | GO:0060537 | muscle tissue development(GO:0060537) |
| 0.0 | 1.0 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.7 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.5 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.6 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.5 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.2 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.0 | 0.1 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.0 | 0.1 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.1 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.3 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 0.9 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 1.3 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.0 | 0.5 | GO:0048332 | mesoderm morphogenesis(GO:0048332) |
| 0.0 | 0.2 | GO:0022011 | Schwann cell differentiation(GO:0014037) Schwann cell development(GO:0014044) myelination in peripheral nervous system(GO:0022011) |
| 0.0 | 0.4 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 1.6 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.2 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0006478 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.2 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.0 | 0.6 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.2 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.2 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.3 | GO:0048546 | digestive tract morphogenesis(GO:0048546) |
| 0.0 | 0.6 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 0.0 | 0.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.3 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.6 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.3 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.0 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.5 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.2 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 1.1 | GO:0002757 | immune response-activating signal transduction(GO:0002757) |
| 0.0 | 0.6 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.8 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 0.2 | GO:0035675 | neuromast hair cell development(GO:0035675) |
| 0.0 | 0.1 | GO:0060538 | skeletal muscle organ development(GO:0060538) |
| 0.0 | 0.2 | GO:0007004 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.0 | 0.2 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.3 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.1 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.0 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.0 | 0.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) negative regulation of gene silencing(GO:0060969) |
| 0.0 | 1.7 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.0 | 0.8 | GO:0001503 | ossification(GO:0001503) |
| 0.0 | 0.2 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.8 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.7 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.1 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 6.9 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.3 | 1.8 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.3 | 1.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.2 | 0.7 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.2 | 0.9 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.2 | 0.7 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.2 | 2.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 0.5 | GO:0043218 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.2 | 2.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.2 | 2.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.2 | 2.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.9 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 2.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.4 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.1 | 16.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 5.4 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.9 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 0.7 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 7.0 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 2.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 10.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 4.2 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 2.2 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.8 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 2.2 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.1 | 0.6 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 4.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.7 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 2.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.8 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.4 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 2.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 0.6 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.8 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.6 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 0.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 1.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.4 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.1 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 3.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 6.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 1.0 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 5.4 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.1 | 0.7 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.3 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 0.2 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.1 | 4.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 6.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 3.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 3.5 | GO:0022626 | cytosolic large ribosomal subunit(GO:0022625) cytosolic ribosome(GO:0022626) |
| 0.1 | 1.5 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 0.2 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.1 | 0.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 2.1 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 5.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.6 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.0 | 0.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.2 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.0 | 0.5 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.8 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0008352 | katanin complex(GO:0008352) |
| 0.0 | 3.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.5 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.7 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.0 | 0.1 | GO:0005775 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.0 | 0.2 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.2 | GO:0030428 | cell septum(GO:0030428) |
| 0.0 | 0.1 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 1.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 2.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.9 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.0 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 1.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 4.7 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 2.4 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.3 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.0 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.4 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.0 | GO:0030055 | cell-substrate junction(GO:0030055) |
| 0.0 | 0.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 3.5 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 13.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 1.3 | 5.0 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 1.1 | 3.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.7 | 2.9 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.5 | 2.7 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.4 | 1.2 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.4 | 3.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.4 | 1.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.3 | 6.9 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.3 | 1.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.3 | 16.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.3 | 1.9 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.3 | 1.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.3 | 1.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.3 | 2.7 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 5.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 2.4 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.2 | 0.7 | GO:1990715 | mRNA CDS binding(GO:1990715) sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 1.9 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.2 | 7.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.2 | 0.7 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 8.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 3.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 0.6 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.2 | 1.0 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.2 | 0.6 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.2 | 0.6 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.2 | 2.8 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.2 | 0.9 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.2 | 1.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.2 | 6.1 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.2 | 0.7 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.2 | 1.7 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.2 | 0.6 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.2 | 1.6 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 0.9 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.2 | 1.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 1.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.2 | 0.6 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.2 | 0.5 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.2 | 0.5 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.2 | 2.2 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.2 | 1.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 1.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.4 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.1 | 1.2 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.4 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 1.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.4 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.1 | 0.8 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 1.3 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.1 | 2.6 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.5 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 0.7 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 0.5 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.1 | 0.7 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 1.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.6 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.4 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.1 | 0.3 | GO:0043185 | vascular endothelial growth factor receptor 3 binding(GO:0043185) |
| 0.1 | 1.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.3 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.1 | 6.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 2.5 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.9 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.3 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.1 | 3.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 1.0 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 1.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 3.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.3 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.1 | 0.8 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.2 | GO:0016496 | substance P receptor activity(GO:0016496) |
| 0.1 | 1.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 4.8 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 1.0 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 2.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 2.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 1.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.3 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.1 | 1.5 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 1.3 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.9 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 1.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.4 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 0.7 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.2 | GO:0032138 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.1 | 0.4 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.8 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.8 | GO:0022884 | protein transmembrane transporter activity(GO:0008320) macromolecule transmembrane transporter activity(GO:0022884) |
| 0.1 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 1.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 1.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.4 | GO:0015157 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.9 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 4.8 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 0.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 6.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.0 | 0.8 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.8 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 1.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.0 | 1.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.9 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.4 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.2 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.4 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.2 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.0 | 1.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.0 | 0.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 2.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 4.3 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.6 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.0 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 2.3 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.5 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.0 | 1.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0052725 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) |
| 0.0 | 0.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.7 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.6 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 2.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.3 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.5 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.0 | 0.4 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 2.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.0 | 1.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.4 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 1.6 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.2 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.0 | 0.7 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.3 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 2.2 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.0 | 0.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 1.0 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.2 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 1.3 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 3.7 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.9 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.0 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.4 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.0 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.1 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.2 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.3 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 6.0 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.2 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.8 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 1.1 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.5 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 6.0 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.1 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.3 | 10.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 0.7 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.2 | 6.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 0.6 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.2 | 6.6 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.2 | 1.0 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 5.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 0.7 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 1.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 0.2 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 0.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 0.9 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 12.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 0.9 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 1.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.7 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 0.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 0.5 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 0.5 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 9.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 0.9 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 1.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 2.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.0 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.4 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 1.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.2 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.5 | 6.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.3 | 3.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.3 | 1.4 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.2 | 10.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 6.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.2 | 5.3 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 4.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 0.8 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 0.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 2.8 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 2.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 5.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.9 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 0.8 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 0.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 0.9 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 0.4 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 1.1 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.1 | 1.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 0.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 4.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 2.8 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.1 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 1.0 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 0.9 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 5.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.2 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 2.2 | REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | Genes involved in MyD88:Mal cascade initiated on plasma membrane |
| 0.0 | 3.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.7 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 1.3 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.0 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.3 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |