Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
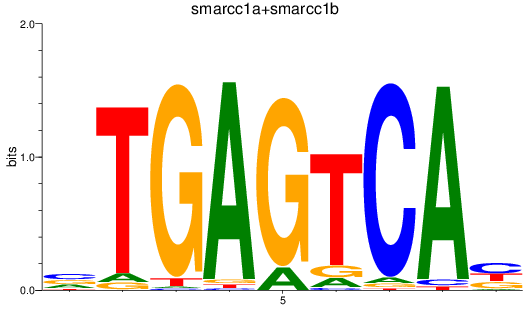
Results for smarcc1a+smarcc1b
Z-value: 1.04
Transcription factors associated with smarcc1a+smarcc1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
smarcc1a
|
ENSDARG00000017397 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1a |
|
smarcc1b
|
ENSDARG00000098919 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| smarcc1b | dr11_v1_chr19_-_19379084_19379084 | 0.08 | 4.7e-01 | Click! |
| smarcc1a | dr11_v1_chr16_-_42461263_42461263 | 0.07 | 5.3e-01 | Click! |
Activity profile of smarcc1a+smarcc1b motif
Sorted Z-values of smarcc1a+smarcc1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_61185746 | 16.20 |
ENSDART00000028219
|
pvalb4
|
parvalbumin 4 |
| chr3_-_32818607 | 12.31 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr22_-_29191152 | 10.75 |
ENSDART00000132702
|
pvalb7
|
parvalbumin 7 |
| chr3_-_4303262 | 9.90 |
ENSDART00000112819
|
si:dkey-73p2.2
|
si:dkey-73p2.2 |
| chr13_+_22479988 | 8.94 |
ENSDART00000188182
ENSDART00000192972 ENSDART00000178372 |
ldb3a
|
LIM domain binding 3a |
| chr23_+_20408227 | 8.65 |
ENSDART00000134727
|
si:rp71-17i16.4
|
si:rp71-17i16.4 |
| chr7_+_7048245 | 8.26 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr19_-_21832441 | 8.20 |
ENSDART00000151272
ENSDART00000151442 ENSDART00000150168 ENSDART00000148797 ENSDART00000128196 ENSDART00000149259 ENSDART00000052556 ENSDART00000149658 ENSDART00000149639 ENSDART00000148424 |
mbpa
|
myelin basic protein a |
| chr1_-_59252973 | 7.41 |
ENSDART00000167061
|
si:ch1073-286c18.5
|
si:ch1073-286c18.5 |
| chr17_-_5583345 | 7.14 |
ENSDART00000035944
|
clic5a
|
chloride intracellular channel 5a |
| chr14_+_30279391 | 7.08 |
ENSDART00000172794
|
fgl1
|
fibrinogen-like 1 |
| chr16_+_23403602 | 7.06 |
ENSDART00000159848
|
s100w
|
S100 calcium binding protein W |
| chr4_+_77948970 | 6.95 |
ENSDART00000149636
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr19_-_38539670 | 6.49 |
ENSDART00000136775
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr22_+_11756040 | 6.23 |
ENSDART00000105808
|
krt97
|
keratin 97 |
| chr13_+_22480496 | 6.05 |
ENSDART00000136863
ENSDART00000131870 ENSDART00000078720 ENSDART00000078740 ENSDART00000139218 |
ldb3a
|
LIM domain binding 3a |
| chr16_-_22303130 | 6.01 |
ENSDART00000142181
|
si:dkey-92i15.4
|
si:dkey-92i15.4 |
| chr2_+_30379650 | 5.93 |
ENSDART00000129542
|
crispld1b
|
cysteine-rich secretory protein LCCL domain containing 1b |
| chr13_-_7031033 | 5.83 |
ENSDART00000193211
|
CABZ01061524.1
|
|
| chr11_-_24681292 | 5.74 |
ENSDART00000089601
|
olfml3b
|
olfactomedin-like 3b |
| chr21_+_25765734 | 5.54 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr12_-_26064480 | 5.48 |
ENSDART00000158215
ENSDART00000171206 ENSDART00000171212 ENSDART00000182956 ENSDART00000186779 |
ldb3b
|
LIM domain binding 3b |
| chr22_-_26595027 | 5.35 |
ENSDART00000184162
|
CABZ01072309.1
|
|
| chr20_-_35578435 | 5.24 |
ENSDART00000142444
|
adgrf6
|
adhesion G protein-coupled receptor F6 |
| chr22_-_24297510 | 5.22 |
ENSDART00000163297
|
si:ch211-117l17.6
|
si:ch211-117l17.6 |
| chr9_-_48281941 | 5.20 |
ENSDART00000099787
|
klhl41a
|
kelch-like family member 41a |
| chr22_+_19247255 | 5.13 |
ENSDART00000144053
|
si:dkey-21e2.10
|
si:dkey-21e2.10 |
| chr14_-_17588345 | 4.94 |
ENSDART00000143486
|
selenot2
|
selenoprotein T, 2 |
| chr6_-_8244474 | 4.86 |
ENSDART00000151358
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr2_+_25278107 | 4.84 |
ENSDART00000131977
|
ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr11_-_18253111 | 4.80 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr13_+_43247936 | 4.62 |
ENSDART00000126850
ENSDART00000165331 |
smoc2
|
SPARC related modular calcium binding 2 |
| chr9_-_9282519 | 4.59 |
ENSDART00000146821
|
si:ch211-214p13.3
|
si:ch211-214p13.3 |
| chr13_+_24842857 | 4.58 |
ENSDART00000123866
|
dusp13a
|
dual specificity phosphatase 13a |
| chr19_-_5332784 | 4.52 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr16_-_14397003 | 4.51 |
ENSDART00000170957
|
crabp2a
|
cellular retinoic acid binding protein 2, a |
| chr8_+_50534948 | 4.43 |
ENSDART00000174435
|
PEBP4
|
phosphatidylethanolamine binding protein 4 |
| chr3_+_4346854 | 4.39 |
ENSDART00000004273
|
si:dkey-73p2.3
|
si:dkey-73p2.3 |
| chr11_-_24458786 | 4.37 |
ENSDART00000089713
|
mxra8a
|
matrix-remodelling associated 8a |
| chr20_-_47347962 | 4.37 |
ENSDART00000080863
|
dtnba
|
dystrobrevin, beta a |
| chr7_+_17992455 | 4.31 |
ENSDART00000089211
ENSDART00000190881 |
ehbp1l1a
|
EH domain binding protein 1-like 1a |
| chr15_+_46329149 | 4.30 |
ENSDART00000128404
|
si:ch1073-340i21.3
|
si:ch1073-340i21.3 |
| chr8_+_27807266 | 4.19 |
ENSDART00000170037
|
capza1b
|
capping protein (actin filament) muscle Z-line, alpha 1b |
| chr23_-_39636195 | 4.17 |
ENSDART00000144439
|
vwa1
|
von Willebrand factor A domain containing 1 |
| chr3_-_27646070 | 4.17 |
ENSDART00000122031
ENSDART00000151027 |
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr20_-_47348116 | 4.11 |
ENSDART00000162087
ENSDART00000160769 ENSDART00000164484 |
dtnba
|
dystrobrevin, beta a |
| chr2_+_9821757 | 4.10 |
ENSDART00000018408
ENSDART00000141227 ENSDART00000144681 ENSDART00000148227 |
anxa13l
|
annexin A13, like |
| chr22_+_19188809 | 4.06 |
ENSDART00000134791
ENSDART00000133682 |
si:dkey-21e2.8
|
si:dkey-21e2.8 |
| chr6_-_40842768 | 4.01 |
ENSDART00000076160
|
mustn1a
|
musculoskeletal, embryonic nuclear protein 1a |
| chr18_-_20869175 | 4.01 |
ENSDART00000090079
|
synm
|
synemin, intermediate filament protein |
| chr4_-_16412084 | 4.00 |
ENSDART00000188460
|
dcn
|
decorin |
| chr21_+_20383837 | 3.98 |
ENSDART00000026430
|
hspb11
|
heat shock protein, alpha-crystallin-related, b11 |
| chr2_-_985417 | 3.82 |
ENSDART00000140540
|
si:ch211-241e1.3
|
si:ch211-241e1.3 |
| chr15_-_2657508 | 3.69 |
ENSDART00000102086
|
cldna
|
claudin a |
| chr10_-_22095505 | 3.65 |
ENSDART00000140210
|
ponzr10
|
plac8 onzin related protein 10 |
| chr20_+_7084154 | 3.60 |
ENSDART00000136448
|
ftr85
|
finTRIM family, member 85 |
| chr11_-_45171139 | 3.58 |
ENSDART00000167036
ENSDART00000161712 ENSDART00000158156 |
syngr2b
|
synaptogyrin 2b |
| chr2_+_9822319 | 3.49 |
ENSDART00000144078
ENSDART00000144371 |
anxa13l
|
annexin A13, like |
| chr3_-_48259289 | 3.47 |
ENSDART00000160717
|
znf750
|
zinc finger protein 750 |
| chr24_+_13869092 | 3.47 |
ENSDART00000176492
|
eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr2_+_36015049 | 3.46 |
ENSDART00000158276
|
lamc2
|
laminin, gamma 2 |
| chr19_-_31035155 | 3.43 |
ENSDART00000161882
|
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr7_+_34549198 | 3.39 |
ENSDART00000173784
|
fhod1
|
formin homology 2 domain containing 1 |
| chr5_-_25582721 | 3.34 |
ENSDART00000123986
|
anxa1a
|
annexin A1a |
| chr7_-_49892991 | 3.32 |
ENSDART00000126240
|
cd44a
|
CD44 molecule (Indian blood group) a |
| chr8_-_44463985 | 3.31 |
ENSDART00000016845
|
mhc1lba
|
major histocompatibility complex class I LBA |
| chr15_-_33834577 | 3.30 |
ENSDART00000163354
|
mmp13b
|
matrix metallopeptidase 13b |
| chr12_-_37299646 | 3.29 |
ENSDART00000146142
ENSDART00000085201 |
pmp22b
|
peripheral myelin protein 22b |
| chr21_+_10866421 | 3.24 |
ENSDART00000137858
|
alpk2
|
alpha-kinase 2 |
| chr5_-_30615901 | 3.14 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr22_+_22888 | 3.13 |
ENSDART00000082471
|
mfap2
|
microfibril associated protein 2 |
| chr6_+_51713076 | 3.12 |
ENSDART00000146281
|
ripor3
|
RIPOR family member 3 |
| chr12_-_4388704 | 3.12 |
ENSDART00000152168
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr19_-_31035325 | 3.11 |
ENSDART00000147504
|
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr8_+_39663612 | 3.10 |
ENSDART00000188074
|
BX005328.1
|
|
| chr4_-_4261673 | 3.09 |
ENSDART00000150694
|
cd9b
|
CD9 molecule b |
| chr7_-_35432901 | 3.07 |
ENSDART00000026712
|
mmp2
|
matrix metallopeptidase 2 |
| chr2_+_55984788 | 3.06 |
ENSDART00000183599
|
nmrk2
|
nicotinamide riboside kinase 2 |
| chr16_+_49005321 | 3.04 |
ENSDART00000160919
|
CABZ01068499.1
|
|
| chr11_-_43104475 | 3.00 |
ENSDART00000125368
|
acyp2
|
acylphosphatase 2, muscle type |
| chr22_+_11775269 | 2.99 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr7_-_71389375 | 2.94 |
ENSDART00000128928
|
CABZ01074298.1
|
|
| chr7_-_22941472 | 2.91 |
ENSDART00000190334
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr4_+_5317483 | 2.85 |
ENSDART00000150366
|
si:ch211-214j24.10
|
si:ch211-214j24.10 |
| chr8_-_23573084 | 2.85 |
ENSDART00000139084
|
wasb
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia) b |
| chr21_-_22828593 | 2.81 |
ENSDART00000150993
|
angptl5
|
angiopoietin-like 5 |
| chr24_-_24797455 | 2.79 |
ENSDART00000138741
|
pde7a
|
phosphodiesterase 7A |
| chr4_+_76671012 | 2.78 |
ENSDART00000005585
|
ms4a17a.2
|
membrane-spanning 4-domains, subfamily A, member 17a.2 |
| chr19_-_7420867 | 2.75 |
ENSDART00000081741
|
rab25a
|
RAB25, member RAS oncogene family a |
| chr16_+_13818500 | 2.72 |
ENSDART00000135245
|
flcn
|
folliculin |
| chr20_+_39283849 | 2.72 |
ENSDART00000002481
ENSDART00000146683 |
scara3
|
scavenger receptor class A, member 3 |
| chr1_+_57235896 | 2.71 |
ENSDART00000152621
|
si:dkey-27j5.7
|
si:dkey-27j5.7 |
| chr3_-_58831683 | 2.69 |
ENSDART00000110292
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr5_-_40297003 | 2.65 |
ENSDART00000097526
|
hspb3
|
heat shock protein, alpha-crystallin-related, b3 |
| chr3_-_32169754 | 2.65 |
ENSDART00000179010
|
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr8_+_27807974 | 2.63 |
ENSDART00000078509
|
capza1b
|
capping protein (actin filament) muscle Z-line, alpha 1b |
| chr8_+_3530761 | 2.60 |
ENSDART00000081272
|
gcn1
|
GCN1 eIF2 alpha kinase activator homolog |
| chr10_+_38610741 | 2.57 |
ENSDART00000126444
|
mmp13a
|
matrix metallopeptidase 13a |
| chr13_+_24280380 | 2.51 |
ENSDART00000184115
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr15_-_41438821 | 2.51 |
ENSDART00000136952
|
nlrc8
|
NLR family CARD domain containing 8 |
| chr16_+_21790870 | 2.51 |
ENSDART00000155039
|
trim108
|
tripartite motif containing 108 |
| chr5_-_48260145 | 2.47 |
ENSDART00000044083
ENSDART00000163250 ENSDART00000135911 |
mef2cb
|
myocyte enhancer factor 2cb |
| chr20_-_13660600 | 2.47 |
ENSDART00000063826
|
TAGAP
|
si:ch211-122h15.4 |
| chr24_+_25913162 | 2.41 |
ENSDART00000143099
ENSDART00000184814 |
map3k15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr22_-_38621438 | 2.40 |
ENSDART00000098330
|
nppc
|
natriuretic peptide C |
| chr3_-_34027178 | 2.40 |
ENSDART00000170201
ENSDART00000151408 |
ighv1-4
ighv14-1
|
immunoglobulin heavy variable 1-4 immunoglobulin heavy variable 14-1 |
| chr1_+_40308077 | 2.38 |
ENSDART00000138992
|
vwa10.2
|
von Willebrand factor A domain containing 10, tandem duplicate 2 |
| chr4_+_76775837 | 2.37 |
ENSDART00000174167
|
ms4a17a.10
|
membrane-spanning 4-domains, subfamily A, member 17A.10 |
| chr25_+_29474583 | 2.37 |
ENSDART00000191189
|
il17rel
|
interleukin 17 receptor E-like |
| chr12_-_43982343 | 2.33 |
ENSDART00000161539
|
CR385054.1
|
|
| chr5_+_6670945 | 2.32 |
ENSDART00000185686
|
pxna
|
paxillin a |
| chr21_-_30254185 | 2.31 |
ENSDART00000101054
|
dnajc18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr17_+_12942021 | 2.31 |
ENSDART00000192514
|
nfkbiab
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha b |
| chr13_-_21701323 | 2.30 |
ENSDART00000164112
|
si:dkey-191g9.7
|
si:dkey-191g9.7 |
| chr15_-_25571865 | 2.30 |
ENSDART00000077836
|
mmp20b
|
matrix metallopeptidase 20b (enamelysin) |
| chr7_+_22657566 | 2.28 |
ENSDART00000141048
|
ponzr5
|
plac8 onzin related protein 5 |
| chr22_-_13350240 | 2.27 |
ENSDART00000154095
ENSDART00000155118 |
si:ch211-227m13.1
|
si:ch211-227m13.1 |
| chr19_-_10881486 | 2.27 |
ENSDART00000168852
ENSDART00000160438 |
PSMD4 (1 of many)
psmd4a
|
proteasome 26S subunit, non-ATPase 4 proteasome 26S subunit, non-ATPase 4a |
| chr24_+_42074143 | 2.26 |
ENSDART00000170514
|
top1mt
|
DNA topoisomerase I mitochondrial |
| chr25_+_29474982 | 2.25 |
ENSDART00000130410
|
il17rel
|
interleukin 17 receptor E-like |
| chr7_+_20512419 | 2.24 |
ENSDART00000173907
|
si:dkey-19b23.14
|
si:dkey-19b23.14 |
| chr25_-_22191733 | 2.24 |
ENSDART00000067478
|
pkp3a
|
plakophilin 3a |
| chr3_+_4266289 | 2.24 |
ENSDART00000101636
|
si:dkey-73p2.1
|
si:dkey-73p2.1 |
| chr4_-_18211 | 2.23 |
ENSDART00000171737
|
ptpn12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr8_+_13389115 | 2.23 |
ENSDART00000184428
ENSDART00000154266 ENSDART00000049469 |
jak3
|
Janus kinase 3 (a protein tyrosine kinase, leukocyte) |
| chr24_+_38201089 | 2.23 |
ENSDART00000132338
|
igl3v2
|
immunoglobulin light 3 variable 2 |
| chr1_-_5746030 | 2.20 |
ENSDART00000150863
|
nrp2a
|
neuropilin 2a |
| chr7_-_20241346 | 2.20 |
ENSDART00000173619
ENSDART00000127699 |
si:ch73-335l21.4
|
si:ch73-335l21.4 |
| chr23_+_19213472 | 2.18 |
ENSDART00000185985
|
apobec2b
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2b |
| chr4_+_76659013 | 2.17 |
ENSDART00000147908
ENSDART00000134229 |
ms4a17a.5
|
membrane-spanning 4-domains, subfamily A, member 17A.5 |
| chr7_+_34506937 | 2.16 |
ENSDART00000111303
|
rfx7a
|
regulatory factor X7a |
| chr17_+_41302660 | 2.16 |
ENSDART00000059480
|
fosl2
|
fos-like antigen 2 |
| chr23_-_36316352 | 2.15 |
ENSDART00000014840
|
nfe2
|
nuclear factor, erythroid 2 |
| chr4_+_77933084 | 2.15 |
ENSDART00000148728
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr9_+_2499627 | 2.14 |
ENSDART00000160782
|
wipf1a
|
WAS/WASL interacting protein family, member 1a |
| chr7_-_26571994 | 2.11 |
ENSDART00000128801
|
si:dkey-62k3.6
|
si:dkey-62k3.6 |
| chr3_+_4113551 | 2.10 |
ENSDART00000192309
|
LO018551.1
|
|
| chr20_-_39273505 | 2.08 |
ENSDART00000153114
|
clu
|
clusterin |
| chr24_+_35947077 | 2.08 |
ENSDART00000173406
|
obscnb
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF b |
| chr25_-_22187397 | 2.07 |
ENSDART00000123211
ENSDART00000139110 |
pkp3a
|
plakophilin 3a |
| chr3_+_27027781 | 2.05 |
ENSDART00000065495
|
emp2
|
epithelial membrane protein 2 |
| chr6_+_18298444 | 2.04 |
ENSDART00000166018
|
card14
|
caspase recruitment domain family, member 14 |
| chr20_+_23501535 | 2.03 |
ENSDART00000177922
ENSDART00000058532 |
palld
|
palladin, cytoskeletal associated protein |
| chr20_-_13660767 | 2.03 |
ENSDART00000127654
|
TAGAP
|
si:ch211-122h15.4 |
| chr23_-_35069805 | 2.02 |
ENSDART00000087219
|
BX294434.1
|
|
| chr3_-_32859335 | 2.02 |
ENSDART00000158916
|
si:dkey-16l2.20
|
si:dkey-16l2.20 |
| chr24_+_38209946 | 2.01 |
ENSDART00000058204
|
CU896602.2
|
|
| chr1_+_54655160 | 2.00 |
ENSDART00000190319
|
si:ch211-202h22.7
|
si:ch211-202h22.7 |
| chr6_-_15757867 | 2.00 |
ENSDART00000063665
|
ackr3b
|
atypical chemokine receptor 3b |
| chr10_-_44482911 | 1.99 |
ENSDART00000085556
|
hip1ra
|
huntingtin interacting protein 1 related a |
| chr20_-_39273987 | 1.99 |
ENSDART00000127173
|
clu
|
clusterin |
| chr7_+_34549377 | 1.96 |
ENSDART00000191814
|
fhod1
|
formin homology 2 domain containing 1 |
| chr8_+_6576940 | 1.96 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr23_-_35064785 | 1.95 |
ENSDART00000172240
|
BX294434.1
|
|
| chr1_+_41849152 | 1.94 |
ENSDART00000053685
|
smox
|
spermine oxidase |
| chr3_-_58650057 | 1.91 |
ENSDART00000057640
|
dhrs7ca
|
dehydrogenase/reductase (SDR family) member 7Ca |
| chr25_-_18140537 | 1.91 |
ENSDART00000113581
|
kitlga
|
kit ligand a |
| chr6_-_35439406 | 1.91 |
ENSDART00000073784
|
rgs5a
|
regulator of G protein signaling 5a |
| chr7_-_20103384 | 1.89 |
ENSDART00000052902
|
acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr10_+_38593645 | 1.88 |
ENSDART00000011573
|
mmp13a
|
matrix metallopeptidase 13a |
| chr25_-_18140305 | 1.87 |
ENSDART00000180222
|
kitlga
|
kit ligand a |
| chr21_-_7035599 | 1.85 |
ENSDART00000139777
|
si:ch211-93g21.1
|
si:ch211-93g21.1 |
| chr13_-_18345854 | 1.85 |
ENSDART00000080107
|
si:dkey-228d14.5
|
si:dkey-228d14.5 |
| chr8_+_39511932 | 1.84 |
ENSDART00000113511
|
lzts1
|
leucine zipper, putative tumor suppressor 1 |
| chr5_+_61556172 | 1.82 |
ENSDART00000131937
|
orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chr24_+_38208823 | 1.79 |
ENSDART00000187538
ENSDART00000190189 |
CU896602.3
|
|
| chr14_+_22022441 | 1.78 |
ENSDART00000149121
|
clcf1
|
cardiotrophin-like cytokine factor 1 |
| chr6_+_52947186 | 1.78 |
ENSDART00000155831
|
uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr11_+_2434262 | 1.78 |
ENSDART00000166445
|
csnk2a4
|
casein kinase 2, alpha 4 polypeptide |
| chr10_+_4499943 | 1.77 |
ENSDART00000125299
|
plk2a
|
polo-like kinase 2a (Drosophila) |
| chr5_+_37785152 | 1.77 |
ENSDART00000053511
ENSDART00000189812 |
myo1ca
|
myosin Ic, paralog a |
| chr1_+_58370526 | 1.77 |
ENSDART00000067775
|
slc27a1b
|
solute carrier family 27 (fatty acid transporter), member 1b |
| chr14_-_41678749 | 1.76 |
ENSDART00000163039
|
fgfrl1b
|
fibroblast growth factor receptor like 1b |
| chr1_+_2190714 | 1.76 |
ENSDART00000132126
|
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr21_-_11657043 | 1.76 |
ENSDART00000141297
|
cast
|
calpastatin |
| chr22_+_37631234 | 1.76 |
ENSDART00000007346
|
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr12_-_1951233 | 1.76 |
ENSDART00000005676
ENSDART00000127937 |
sox9a
|
SRY (sex determining region Y)-box 9a |
| chr4_+_76787274 | 1.75 |
ENSDART00000156344
|
si:ch73-56d11.3
|
si:ch73-56d11.3 |
| chr10_-_74408 | 1.75 |
ENSDART00000100073
ENSDART00000141723 |
dyrk1aa
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, a |
| chr23_-_270847 | 1.74 |
ENSDART00000191867
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr8_-_18667693 | 1.74 |
ENSDART00000100516
|
stap2b
|
signal transducing adaptor family member 2b |
| chr16_-_51288178 | 1.73 |
ENSDART00000079864
|
zgc:173729
|
zgc:173729 |
| chr15_+_20352123 | 1.72 |
ENSDART00000011030
ENSDART00000163532 ENSDART00000169537 ENSDART00000161047 |
il15l
|
interleukin 15, like |
| chr24_+_42131564 | 1.71 |
ENSDART00000153854
|
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr24_+_38216340 | 1.70 |
ENSDART00000188561
|
CU896602.4
|
|
| chr18_+_47313715 | 1.70 |
ENSDART00000138806
|
barx2
|
BARX homeobox 2 |
| chr6_+_56141852 | 1.69 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr16_+_53125918 | 1.69 |
ENSDART00000102170
|
CABZ01053976.1
|
|
| chr12_+_22576404 | 1.68 |
ENSDART00000172053
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr9_-_42418470 | 1.68 |
ENSDART00000144353
|
calcrla
|
calcitonin receptor-like a |
| chr19_+_43314700 | 1.66 |
ENSDART00000049045
ENSDART00000133158 |
matn1
|
matrilin 1 |
| chr11_+_45255774 | 1.65 |
ENSDART00000172838
|
aspscr1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr1_+_9004719 | 1.64 |
ENSDART00000006211
ENSDART00000137211 |
prkcba
|
protein kinase C, beta a |
| chr25_-_33275666 | 1.64 |
ENSDART00000193605
|
CABZ01046954.1
|
|
| chr8_-_22288258 | 1.63 |
ENSDART00000140978
ENSDART00000100046 |
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr16_+_23960744 | 1.61 |
ENSDART00000058965
|
apoeb
|
apolipoprotein Eb |
| chr16_+_41517188 | 1.61 |
ENSDART00000049976
|
si:dkey-11p23.7
|
si:dkey-11p23.7 |
| chr17_-_30635298 | 1.60 |
ENSDART00000155478
|
sh3yl1
|
SH3 and SYLF domain containing 1 |
| chr7_+_22688781 | 1.59 |
ENSDART00000173509
|
ugt5g1
|
UDP glucuronosyltransferase 5 family, polypeptide G1 |
| chr24_+_38223618 | 1.59 |
ENSDART00000003495
|
igl3v2
|
immunoglobulin light 3 variable 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of smarcc1a+smarcc1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 8.8 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.9 | 9.1 | GO:0001765 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.8 | 3.2 | GO:0048914 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.7 | 2.2 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.7 | 2.2 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.7 | 2.0 | GO:0003071 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 0.6 | 1.7 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.6 | 3.3 | GO:0031394 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.5 | 1.6 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.5 | 3.6 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.5 | 3.0 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.5 | 2.0 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) |
| 0.5 | 3.3 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.4 | 5.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.4 | 3.5 | GO:2001239 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.4 | 1.5 | GO:0033153 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.4 | 0.8 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.4 | 2.6 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.4 | 6.0 | GO:0050927 | regulation of positive chemotaxis(GO:0050926) positive regulation of positive chemotaxis(GO:0050927) induction of positive chemotaxis(GO:0050930) |
| 0.4 | 1.8 | GO:0031652 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.3 | 1.0 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.3 | 10.8 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.3 | 1.6 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.3 | 4.0 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.3 | 1.2 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.3 | 1.8 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.3 | 2.9 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.3 | 4.3 | GO:0048385 | regulation of retinoic acid receptor signaling pathway(GO:0048385) |
| 0.3 | 5.2 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.3 | 1.3 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.3 | 1.0 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.3 | 1.8 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.2 | 1.0 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.2 | 2.5 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.2 | 1.7 | GO:0042102 | positive regulation of T cell proliferation(GO:0042102) |
| 0.2 | 5.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.2 | 8.7 | GO:1903038 | negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.2 | 1.5 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.2 | 1.5 | GO:0060420 | regulation of heart growth(GO:0060420) |
| 0.2 | 2.2 | GO:0045661 | regulation of myoblast differentiation(GO:0045661) |
| 0.2 | 0.6 | GO:0003249 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.2 | 4.0 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.2 | 2.5 | GO:0090279 | regulation of calcium ion import(GO:0090279) |
| 0.2 | 1.0 | GO:0006031 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.2 | 0.8 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.2 | 1.2 | GO:0035093 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.2 | 8.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 2.8 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.2 | 0.9 | GO:0046620 | ventriculo bulbo valve development(GO:0003173) regulation of organ growth(GO:0046620) |
| 0.2 | 0.9 | GO:0060855 | lymphatic endothelial cell differentiation(GO:0060836) venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.2 | 0.6 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.2 | 0.7 | GO:0003242 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.2 | 0.9 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.2 | 2.6 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.2 | 7.6 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.2 | 4.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.2 | 8.1 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.2 | 1.3 | GO:0044259 | collagen metabolic process(GO:0032963) multicellular organismal macromolecule metabolic process(GO:0044259) |
| 0.2 | 4.6 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.2 | 0.3 | GO:0002676 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.2 | 0.6 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.2 | 1.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.2 | 2.3 | GO:0061055 | myotome development(GO:0061055) |
| 0.2 | 1.4 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 1.3 | GO:0071376 | neutrophil homeostasis(GO:0001780) response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.4 | GO:0098725 | somatic stem cell population maintenance(GO:0035019) symmetric cell division(GO:0098725) |
| 0.1 | 2.2 | GO:0038084 | vascular endothelial growth factor signaling pathway(GO:0038084) |
| 0.1 | 4.0 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 4.6 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 2.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 1.2 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 2.0 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 4.4 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.1 | 1.0 | GO:0031179 | peptide modification(GO:0031179) |
| 0.1 | 1.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 1.0 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.1 | 1.7 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.1 | 5.7 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.1 | 0.4 | GO:0003222 | ventricular trabecula myocardium morphogenesis(GO:0003222) |
| 0.1 | 15.5 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.1 | 4.0 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 0.9 | GO:0043363 | nucleate erythrocyte differentiation(GO:0043363) |
| 0.1 | 2.8 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 0.9 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 0.7 | GO:0050655 | dermatan sulfate proteoglycan metabolic process(GO:0050655) |
| 0.1 | 1.8 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.1 | 2.6 | GO:0071218 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.1 | 4.8 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 0.7 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.4 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.1 | 0.5 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.1 | 1.5 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 0.6 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 3.9 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.1 | 1.1 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 1.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.9 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.5 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.1 | 0.5 | GO:0090134 | coronary vasculature development(GO:0060976) mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.1 | 0.3 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 3.5 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.1 | 0.6 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 2.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 2.5 | GO:0097202 | activation of cysteine-type endopeptidase activity(GO:0097202) |
| 0.1 | 1.1 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 1.4 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 1.8 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.6 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.8 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.2 | GO:0048660 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.1 | 3.3 | GO:1901214 | regulation of neuron death(GO:1901214) |
| 0.1 | 1.5 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.1 | 0.5 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.1 | 0.5 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 1.5 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 3.8 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.1 | 0.7 | GO:0019372 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.3 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 1.8 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.1 | 0.7 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.1 | 3.5 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 4.0 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.1 | 3.8 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 1.7 | GO:0060840 | artery development(GO:0060840) |
| 0.1 | 1.0 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 2.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 1.0 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.1 | 0.2 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 3.1 | GO:0048048 | embryonic eye morphogenesis(GO:0048048) |
| 0.1 | 3.3 | GO:0007338 | single fertilization(GO:0007338) |
| 0.1 | 1.1 | GO:0045104 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.6 | GO:0060113 | mechanoreceptor differentiation(GO:0042490) inner ear receptor cell differentiation(GO:0060113) |
| 0.0 | 2.5 | GO:0014904 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 1.1 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.4 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 1.2 | GO:0046514 | ceramide catabolic process(GO:0046514) |
| 0.0 | 1.1 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 1.1 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 1.5 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 2.3 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 1.6 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.9 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.4 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 1.1 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.5 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 0.2 | GO:0031054 | production of siRNA involved in RNA interference(GO:0030422) primary miRNA processing(GO:0031053) pre-miRNA processing(GO:0031054) |
| 0.0 | 0.8 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.6 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.8 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.2 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.5 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.9 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 1.2 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 0.9 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.6 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 4.2 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 3.7 | GO:0008544 | epidermis development(GO:0008544) |
| 0.0 | 0.2 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.4 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 1.3 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 0.7 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 1.4 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 1.7 | GO:0050871 | phagocytosis, recognition(GO:0006910) positive regulation of B cell activation(GO:0050871) |
| 0.0 | 0.6 | GO:0007164 | establishment of planar polarity(GO:0001736) establishment of tissue polarity(GO:0007164) |
| 0.0 | 2.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.5 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.3 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.0 | 2.0 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
| 0.0 | 6.2 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0070307 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.4 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 3.0 | GO:0042176 | regulation of protein catabolic process(GO:0042176) |
| 0.0 | 0.2 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.2 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.3 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 0.9 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 2.3 | GO:0045216 | cell-cell junction organization(GO:0045216) |
| 0.0 | 1.4 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.4 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 1.3 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 1.7 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.0 | 0.4 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.5 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.0 | 0.7 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.5 | GO:0016441 | posttranscriptional gene silencing(GO:0016441) posttranscriptional gene silencing by RNA(GO:0035194) |
| 0.0 | 0.6 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.2 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.4 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.2 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 0.5 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 6.8 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 1.1 | 3.3 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.6 | 6.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.6 | 2.2 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.5 | 22.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.5 | 8.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.4 | 4.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.4 | 1.6 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.3 | 1.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.3 | 1.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.3 | 2.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.3 | 1.4 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.3 | 11.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.3 | 1.0 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 10.7 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.2 | 3.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.2 | 5.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 0.8 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.2 | 18.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 1.9 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.2 | 2.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.8 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 4.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.0 | GO:0030428 | cell septum(GO:0030428) |
| 0.1 | 0.5 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 2.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 3.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.6 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 1.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 11.7 | GO:0044420 | extracellular matrix component(GO:0044420) |
| 0.1 | 2.1 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.9 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 4.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 6.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 10.7 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 22.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 2.2 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 0.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 1.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.5 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 6.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 8.5 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 1.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.2 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.1 | 0.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 4.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 1.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 3.6 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 2.9 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 1.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 2.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.6 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.4 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.5 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.6 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 5.8 | GO:0005912 | adherens junction(GO:0005912) anchoring junction(GO:0070161) |
| 0.0 | 1.4 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 2.7 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.6 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.6 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.4 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 5.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 2.8 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 4.3 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.5 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.4 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 1.2 | GO:0016607 | nuclear speck(GO:0016607) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.0 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 1.1 | 4.4 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.9 | 18.8 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.8 | 3.0 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.6 | 4.5 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.6 | 4.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.6 | 15.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.6 | 1.7 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.6 | 3.3 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.5 | 4.9 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.5 | 8.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.5 | 1.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.5 | 2.6 | GO:0038131 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.5 | 1.5 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 0.5 | 3.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.4 | 2.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.4 | 2.8 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.4 | 1.9 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.4 | 4.6 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.3 | 1.3 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.3 | 1.3 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.3 | 2.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.3 | 1.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.3 | 3.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.3 | 1.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 0.7 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.2 | 2.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.2 | 1.6 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.2 | 1.5 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.2 | 1.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 1.0 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 1.9 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 1.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 2.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 0.5 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.2 | 2.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.2 | 22.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 0.8 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.2 | 1.7 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.2 | 2.3 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.2 | 4.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.7 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.4 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.8 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.6 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 1.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 1.0 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.1 | 5.9 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 1.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 2.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 4.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.3 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 0.3 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.1 | 1.0 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.1 | 1.4 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 1.5 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 2.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.4 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 3.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.9 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 1.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 9.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 1.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.4 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.1 | 0.3 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.1 | 2.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 1.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 2.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 12.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.3 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.1 | 3.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 0.3 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 1.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.2 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 1.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 0.2 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.1 | 1.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 2.5 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 0.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.5 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.1 | 0.8 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.1 | 0.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.6 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 1.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.2 | GO:0072571 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.1 | 0.6 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 0.3 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.1 | 3.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 1.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.2 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 0.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 2.5 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.2 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.0 | 2.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.9 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 1.2 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 1.2 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.6 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 1.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.2 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 4.4 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 1.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.6 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.0 | 36.2 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 11.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 20.9 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.8 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 1.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.5 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.0 | 7.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 2.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.6 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.5 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 1.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.7 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 15.9 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 1.0 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.2 | GO:0051427 | hormone receptor binding(GO:0051427) |
| 0.0 | 2.5 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 1.9 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 4.1 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 8.5 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 3.1 | GO:0004175 | endopeptidase activity(GO:0004175) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 3.3 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 3.9 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 0.8 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 1.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.2 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.5 | GO:0022839 | calcium activated cation channel activity(GO:0005227) ion gated channel activity(GO:0022839) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.5 | GO:0005126 | cytokine receptor binding(GO:0005126) |
| 0.0 | 0.1 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.0 | 0.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.9 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.4 | 3.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.3 | 9.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 6.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 1.3 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.2 | 0.9 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 4.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.8 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 2.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 4.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 4.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 1.5 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 1.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 0.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 1.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 1.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 1.7 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 4.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.7 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.2 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.4 | 1.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.3 | 4.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 8.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.2 | 2.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 1.8 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 1.4 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 2.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 0.9 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 6.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.8 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 1.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 2.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 2.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.7 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 1.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 0.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 3.5 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 0.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 1.0 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 1.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 5.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 1.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.7 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 1.7 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 2.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.7 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 0.2 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |