Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
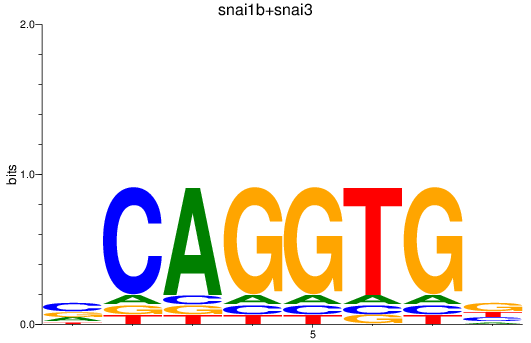
Results for snai1b+snai3
Z-value: 0.58
Transcription factors associated with snai1b+snai3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
snai3
|
ENSDARG00000031243 | snail family zinc finger 3 |
|
snai1b
|
ENSDARG00000046019 | snail family zinc finger 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| snai3 | dr11_v1_chr7_+_55112922_55112922 | 0.20 | 5.1e-02 | Click! |
| snai1b | dr11_v1_chr23_-_2513300_2513300 | -0.14 | 1.8e-01 | Click! |
Activity profile of snai1b+snai3 motif
Sorted Z-values of snai1b+snai3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_32039386 | 4.95 |
ENSDART00000133801
|
mylk2
|
myosin light chain kinase 2 |
| chr13_-_37180815 | 4.17 |
ENSDART00000139907
|
si:dkeyp-77c8.1
|
si:dkeyp-77c8.1 |
| chr12_+_30586599 | 3.53 |
ENSDART00000124920
ENSDART00000126984 |
nrap
|
nebulin-related anchoring protein |
| chr13_+_23157053 | 3.48 |
ENSDART00000162359
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr6_-_10788065 | 2.72 |
ENSDART00000190968
|
wipf1b
|
WAS/WASL interacting protein family, member 1b |
| chr5_+_6670945 | 2.56 |
ENSDART00000185686
|
pxna
|
paxillin a |
| chr14_+_15155684 | 2.51 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr10_+_1052591 | 2.49 |
ENSDART00000123405
|
unc5c
|
unc-5 netrin receptor C |
| chr20_-_9436521 | 2.43 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr4_-_78026285 | 2.35 |
ENSDART00000168273
|
CCT2
|
chaperonin containing TCP1 subunit 2 |
| chr10_-_33588106 | 2.27 |
ENSDART00000155634
|
eva1c
|
eva-1 homolog C (C. elegans) |
| chr21_-_43666420 | 2.14 |
ENSDART00000139008
ENSDART00000183996 ENSDART00000183395 |
si:dkey-229d11.3
si:dkey-229d11.5
|
si:dkey-229d11.3 si:dkey-229d11.5 |
| chr25_+_5249513 | 1.97 |
ENSDART00000126814
|
CABZ01039863.1
|
|
| chr22_-_14128716 | 1.93 |
ENSDART00000140323
|
si:ch211-246m6.4
|
si:ch211-246m6.4 |
| chr20_-_23426339 | 1.92 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr25_-_169291 | 1.88 |
ENSDART00000128344
|
lipcb
|
lipase, hepatic b |
| chr19_-_35439237 | 1.64 |
ENSDART00000145883
|
anln
|
anillin, actin binding protein |
| chr20_-_43743700 | 1.64 |
ENSDART00000100620
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr1_-_55058795 | 1.60 |
ENSDART00000187293
|
peli1a
|
pellino E3 ubiquitin protein ligase 1a |
| chr22_+_20560041 | 1.58 |
ENSDART00000135490
|
nfil3-4
|
nuclear factor, interleukin 3 regulated, member 4 |
| chr7_+_58751504 | 1.53 |
ENSDART00000024185
|
zgc:56231
|
zgc:56231 |
| chr2_-_10896745 | 1.53 |
ENSDART00000114609
|
cdcp2
|
CUB domain containing protein 2 |
| chr14_-_26482096 | 1.45 |
ENSDART00000187280
|
smad5
|
SMAD family member 5 |
| chr10_-_13116337 | 1.44 |
ENSDART00000164568
|
musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr7_-_45990681 | 1.39 |
ENSDART00000165441
|
si:ch211-260e23.7
|
si:ch211-260e23.7 |
| chr1_-_43892349 | 1.39 |
ENSDART00000148416
|
tacr3a
|
tachykinin receptor 3a |
| chr10_+_24445698 | 1.38 |
ENSDART00000146370
|
slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr17_-_11151655 | 1.37 |
ENSDART00000156383
|
CU179699.1
|
|
| chr20_-_21672970 | 1.36 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr23_+_34047413 | 1.36 |
ENSDART00000143933
ENSDART00000123925 ENSDART00000176139 |
chchd6a
|
coiled-coil-helix-coiled-coil-helix domain containing 6a |
| chr1_+_524717 | 1.33 |
ENSDART00000102421
ENSDART00000184473 |
mrpl16
|
mitochondrial ribosomal protein L16 |
| chr20_-_52902693 | 1.29 |
ENSDART00000166115
ENSDART00000161050 |
ctsbb
|
cathepsin Bb |
| chr19_+_142270 | 1.26 |
ENSDART00000160201
|
tnfrsf11b
|
tumor necrosis factor receptor superfamily, member 11b |
| chr2_-_49997055 | 1.21 |
ENSDART00000140294
|
si:ch211-106n13.3
|
si:ch211-106n13.3 |
| chr25_-_16076257 | 1.19 |
ENSDART00000140780
|
ovch2
|
ovochymase 2 |
| chr18_+_33580660 | 1.19 |
ENSDART00000139117
|
si:dkey-47k20.4
|
si:dkey-47k20.4 |
| chr20_-_1314355 | 1.18 |
ENSDART00000152436
|
pcmt
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr22_-_209741 | 1.15 |
ENSDART00000171954
|
ora1
|
olfactory receptor class A related 1 |
| chr4_-_12790886 | 1.14 |
ENSDART00000182535
ENSDART00000067131 ENSDART00000186426 |
irak3
|
interleukin-1 receptor-associated kinase 3 |
| chr18_+_27821856 | 1.13 |
ENSDART00000131712
|
si:ch211-222m18.4
|
si:ch211-222m18.4 |
| chr5_-_22052852 | 1.11 |
ENSDART00000002938
|
mtmr8
|
myotubularin related protein 8 |
| chr21_+_19858627 | 1.10 |
ENSDART00000147010
|
fybb
|
FYN binding protein b |
| chr13_-_36579086 | 1.07 |
ENSDART00000146671
|
lgals3a
|
lectin, galactoside binding soluble 3a |
| chr3_-_34052882 | 1.05 |
ENSDART00000151463
|
ighv11-1
|
immunoglobulin heavy variable 11-1 |
| chr24_+_19578935 | 1.02 |
ENSDART00000137175
|
sulf1
|
sulfatase 1 |
| chr5_-_26181863 | 1.01 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr7_-_50367326 | 0.99 |
ENSDART00000141926
|
prc1b
|
protein regulator of cytokinesis 1b |
| chr13_+_35955562 | 0.92 |
ENSDART00000137377
|
si:ch211-67f13.7
|
si:ch211-67f13.7 |
| chr23_-_29824146 | 0.90 |
ENSDART00000020616
|
zgc:194189
|
zgc:194189 |
| chr22_-_17677947 | 0.87 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr6_+_39905021 | 0.86 |
ENSDART00000064904
|
endou
|
endonuclease, polyU-specific |
| chr9_-_28255029 | 0.86 |
ENSDART00000160387
|
ccnyl1
|
cyclin Y-like 1 |
| chr18_-_37241080 | 0.83 |
ENSDART00000126421
ENSDART00000078064 |
six9
|
SIX homeobox 9 |
| chr24_+_9372292 | 0.77 |
ENSDART00000082422
ENSDART00000191127 ENSDART00000180510 |
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr10_+_37173029 | 0.76 |
ENSDART00000136510
|
ksr1a
|
kinase suppressor of ras 1a |
| chr7_-_50367642 | 0.75 |
ENSDART00000134941
|
prc1b
|
protein regulator of cytokinesis 1b |
| chr8_-_13029297 | 0.75 |
ENSDART00000144305
|
dennd2da
|
DENN/MADD domain containing 2Da |
| chr14_-_38865800 | 0.74 |
ENSDART00000173047
|
gsr
|
glutathione reductase |
| chr7_+_30926289 | 0.74 |
ENSDART00000173518
|
si:dkey-1h4.4
|
si:dkey-1h4.4 |
| chr6_-_130849 | 0.74 |
ENSDART00000108710
|
LRRC8E
|
si:zfos-323e3.4 |
| chr23_-_44219902 | 0.71 |
ENSDART00000185874
|
zgc:158659
|
zgc:158659 |
| chr9_-_11549379 | 0.71 |
ENSDART00000187074
|
fev
|
FEV (ETS oncogene family) |
| chr4_-_16341801 | 0.70 |
ENSDART00000140190
|
kera
|
keratocan |
| chr10_-_8358396 | 0.70 |
ENSDART00000059322
|
csgalnact1a
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1a |
| chr22_+_7439186 | 0.69 |
ENSDART00000190667
|
zgc:92041
|
zgc:92041 |
| chr22_+_22438783 | 0.69 |
ENSDART00000147825
|
kif14
|
kinesin family member 14 |
| chr25_+_5012791 | 0.68 |
ENSDART00000156970
|
si:ch73-265h17.5
|
si:ch73-265h17.5 |
| chr9_-_849069 | 0.68 |
ENSDART00000136091
|
c1ql2
|
complement component 1, q subcomponent-like 2 |
| chr10_-_38468847 | 0.67 |
ENSDART00000133914
|
gdpd5a
|
glycerophosphodiester phosphodiesterase domain containing 5a |
| chr25_-_17395315 | 0.66 |
ENSDART00000064596
|
cyp2x8
|
cytochrome P450, family 2, subfamily X, polypeptide 8 |
| chr14_-_28052474 | 0.66 |
ENSDART00000172948
ENSDART00000135337 |
TSC22D3 (1 of many)
zgc:64189
|
si:ch211-220e11.3 zgc:64189 |
| chr23_-_24047054 | 0.64 |
ENSDART00000184308
ENSDART00000185902 |
CR925720.1
|
|
| chr13_-_9119867 | 0.62 |
ENSDART00000137255
|
si:dkey-112g5.15
|
si:dkey-112g5.15 |
| chr6_-_10728057 | 0.60 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr15_-_434503 | 0.57 |
ENSDART00000122286
|
CABZ01056629.1
|
|
| chr18_+_45666489 | 0.56 |
ENSDART00000180147
ENSDART00000151351 |
prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr8_-_34762163 | 0.56 |
ENSDART00000114080
|
setd1bb
|
SET domain containing 1B, b |
| chr24_-_25184553 | 0.56 |
ENSDART00000166917
|
plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr7_+_42328578 | 0.55 |
ENSDART00000149082
|
phkb
|
phosphorylase kinase, beta |
| chr2_+_5948534 | 0.54 |
ENSDART00000124324
ENSDART00000176461 |
slc1a7a
|
solute carrier family 1 (glutamate transporter), member 7a |
| chr17_+_1360192 | 0.54 |
ENSDART00000184561
|
siva1
|
SIVA1, apoptosis-inducing factor |
| chr9_+_331425 | 0.51 |
ENSDART00000170312
|
actr5
|
ARP5 actin related protein 5 homolog |
| chr3_+_23029934 | 0.50 |
ENSDART00000110343
|
nags
|
N-acetylglutamate synthase |
| chr11_+_6902946 | 0.47 |
ENSDART00000144006
|
crtc1b
|
CREB regulated transcription coactivator 1b |
| chr11_+_40831620 | 0.46 |
ENSDART00000160023
|
errfi1a
|
ERBB receptor feedback inhibitor 1a |
| chr8_+_247163 | 0.44 |
ENSDART00000122378
|
cep120
|
centrosomal protein 120 |
| chr4_-_36476889 | 0.43 |
ENSDART00000163956
|
si:ch211-263l8.1
|
si:ch211-263l8.1 |
| chr1_-_21909608 | 0.42 |
ENSDART00000139937
|
frmpd1a
|
FERM and PDZ domain containing 1a |
| chr15_-_42736433 | 0.42 |
ENSDART00000154379
|
si:ch211-181d7.1
|
si:ch211-181d7.1 |
| chr22_-_8910567 | 0.41 |
ENSDART00000187896
ENSDART00000188511 ENSDART00000175764 |
FO393424.2
|
|
| chr12_-_46112892 | 0.41 |
ENSDART00000187128
ENSDART00000114268 |
zgc:153932
|
zgc:153932 |
| chr2_+_9536021 | 0.40 |
ENSDART00000187357
|
CU929379.1
|
|
| chr20_+_42978499 | 0.39 |
ENSDART00000138793
|
si:ch211-203k16.3
|
si:ch211-203k16.3 |
| chr22_-_18546649 | 0.39 |
ENSDART00000171277
|
cirbpb
|
cold inducible RNA binding protein b |
| chr4_+_54645654 | 0.37 |
ENSDART00000192864
|
si:ch211-227e10.1
|
si:ch211-227e10.1 |
| chr5_+_53482597 | 0.36 |
ENSDART00000180333
|
BX323994.1
|
|
| chr12_+_48784731 | 0.36 |
ENSDART00000158348
|
zmiz1b
|
zinc finger, MIZ-type containing 1b |
| chr21_-_41873584 | 0.30 |
ENSDART00000188089
|
endou2
|
endonuclease, polyU-specific 2 |
| chr13_+_2448251 | 0.23 |
ENSDART00000188361
|
arfgef3
|
ARFGEF family member 3 |
| chr7_-_58867188 | 0.22 |
ENSDART00000187006
|
CU681855.1
|
|
| chr21_+_30794351 | 0.17 |
ENSDART00000139486
|
zgc:158225
|
zgc:158225 |
| chr2_-_42628028 | 0.16 |
ENSDART00000179866
|
myo10
|
myosin X |
| chr3_+_56276346 | 0.15 |
ENSDART00000157770
|
PRKCA
|
si:ch73-374l24.1 |
| chr10_+_9601587 | 0.13 |
ENSDART00000147997
|
rc3h2
|
ring finger and CCCH-type domains 2 |
| chr11_-_8782871 | 0.13 |
ENSDART00000158546
|
si:ch211-51h4.2
|
si:ch211-51h4.2 |
| chr8_-_10949847 | 0.13 |
ENSDART00000123209
|
pqlc2
|
PQ loop repeat containing 2 |
| chr2_-_722156 | 0.11 |
ENSDART00000045770
ENSDART00000169498 |
foxq1a
|
forkhead box Q1a |
| chr10_-_94184 | 0.07 |
ENSDART00000146125
|
ttc3
|
tetratricopeptide repeat domain 3 |
| chr22_-_17256573 | 0.04 |
ENSDART00000136119
ENSDART00000062891 |
nphs2
|
nephrosis 2, idiopathic, steroid-resistant (podocin) |
| chr18_+_33580391 | 0.00 |
ENSDART00000189300
|
si:dkey-47k20.4
|
si:dkey-47k20.4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of snai1b+snai3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.5 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.4 | 1.6 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.4 | 1.1 | GO:0032640 | tumor necrosis factor production(GO:0032640) tumor necrosis factor superfamily cytokine production(GO:0071706) |
| 0.3 | 1.6 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.2 | 3.6 | GO:0061055 | myotome development(GO:0061055) |
| 0.2 | 2.5 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.2 | 1.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.2 | 1.4 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.2 | 1.4 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.2 | 0.9 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.2 | 1.7 | GO:0000022 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.5 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 1.9 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.4 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.1 | 1.1 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.1 | 1.5 | GO:2000223 | regulation of BMP signaling pathway involved in heart jogging(GO:2000223) |
| 0.1 | 0.9 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.1 | 1.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.7 | GO:0071684 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 1.0 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 0.5 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 0.5 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 0.9 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 1.4 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.7 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 1.1 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 1.3 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.7 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 2.6 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.7 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.6 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 1.1 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 1.0 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 1.9 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.7 | GO:0051057 | positive regulation of Ras protein signal transduction(GO:0046579) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.0 | 1.3 | GO:0009612 | response to mechanical stimulus(GO:0009612) |
| 0.0 | 1.6 | GO:0007623 | circadian rhythm(GO:0007623) |
| 0.0 | 0.7 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 1.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 1.7 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 1.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.9 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 1.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.7 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 3.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.5 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.6 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.3 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 2.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 2.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 2.6 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 1.7 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 1.2 | GO:0005581 | collagen trimer(GO:0005581) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.4 | 1.1 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.4 | 5.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 2.5 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.2 | 1.1 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.2 | 1.4 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.2 | 1.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.2 | 1.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 0.7 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.1 | 0.4 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 1.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.7 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 1.5 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.7 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 1.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.9 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 1.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 2.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 2.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.2 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 5.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.5 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 0.7 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 1.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 2.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 1.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 3.5 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 1.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 0.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.1 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |