Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
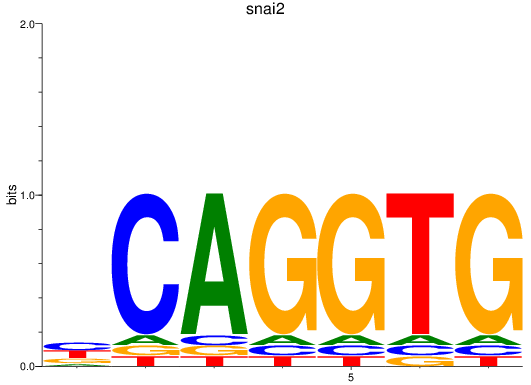
Results for snai2
Z-value: 0.67
Transcription factors associated with snai2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
snai2
|
ENSDARG00000040046 | snail family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| snai2 | dr11_v1_chr24_+_35387517_35387517 | -0.20 | 5.1e-02 | Click! |
Activity profile of snai2 motif
Sorted Z-values of snai2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_106689 | 10.84 |
ENSDART00000185007
|
apoa1b
|
apolipoprotein A-Ib |
| chr20_+_10538025 | 8.74 |
ENSDART00000129762
|
serpina1l
|
serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1, like |
| chr15_+_20239141 | 6.91 |
ENSDART00000101152
ENSDART00000152473 |
spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr22_+_661711 | 6.58 |
ENSDART00000113795
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr22_+_661505 | 6.20 |
ENSDART00000149460
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr21_-_30658509 | 5.98 |
ENSDART00000139764
|
si:dkey-22f5.9
|
si:dkey-22f5.9 |
| chr25_-_17395315 | 5.45 |
ENSDART00000064596
|
cyp2x8
|
cytochrome P450, family 2, subfamily X, polypeptide 8 |
| chr19_-_48312109 | 5.42 |
ENSDART00000161103
|
si:ch73-359m17.9
|
si:ch73-359m17.9 |
| chr7_-_53117131 | 5.11 |
ENSDART00000169211
ENSDART00000168890 ENSDART00000172179 ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr2_-_32643738 | 4.64 |
ENSDART00000112452
|
si:dkeyp-73d8.9
|
si:dkeyp-73d8.9 |
| chr3_-_15080226 | 4.63 |
ENSDART00000109818
ENSDART00000139835 |
nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr7_-_26306546 | 4.63 |
ENSDART00000140817
|
zgc:77439
|
zgc:77439 |
| chr11_+_329687 | 4.62 |
ENSDART00000172882
|
cyp27b1
|
cytochrome P450, family 27, subfamily B, polypeptide 1 |
| chr23_-_12735434 | 4.44 |
ENSDART00000139166
|
si:dkey-96f10.1
|
si:dkey-96f10.1 |
| chr7_+_52122224 | 4.41 |
ENSDART00000174268
|
cyp2x12
|
cytochrome P450, family 2, subfamily X, polypeptide 12 |
| chr2_+_4208323 | 4.00 |
ENSDART00000167906
|
gata6
|
GATA binding protein 6 |
| chr20_-_33556983 | 3.99 |
ENSDART00000168798
|
rock2bl
|
rho-associated, coiled-coil containing protein kinase 2b, like |
| chr14_+_15155684 | 3.94 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr1_+_56180416 | 3.74 |
ENSDART00000089358
|
crb3b
|
crumbs homolog 3b |
| chr20_-_43743700 | 3.54 |
ENSDART00000100620
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr15_+_46606090 | 3.52 |
ENSDART00000020921
|
CABZ01044048.1
|
|
| chr19_+_791538 | 3.34 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr12_-_4243268 | 3.28 |
ENSDART00000131275
|
zgc:92313
|
zgc:92313 |
| chr17_-_49412313 | 3.21 |
ENSDART00000152100
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr18_+_45666489 | 3.20 |
ENSDART00000180147
ENSDART00000151351 |
prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr14_-_14659023 | 3.15 |
ENSDART00000170355
ENSDART00000159888 ENSDART00000172241 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr7_+_14005111 | 3.05 |
ENSDART00000187365
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr20_-_3238110 | 3.05 |
ENSDART00000008077
|
spint1b
|
serine peptidase inhibitor, Kunitz type 1 b |
| chr3_+_1179601 | 2.91 |
ENSDART00000173378
|
triobpb
|
TRIO and F-actin binding protein b |
| chr13_+_37022601 | 2.89 |
ENSDART00000131800
ENSDART00000041300 |
esr2b
|
estrogen receptor 2b |
| chr21_+_45839917 | 2.86 |
ENSDART00000189305
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr4_-_9891874 | 2.86 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr16_+_23961276 | 2.85 |
ENSDART00000192754
|
apoeb
|
apolipoprotein Eb |
| chr5_-_6561376 | 2.83 |
ENSDART00000099570
|
tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr18_+_21061216 | 2.76 |
ENSDART00000141739
|
fam169b
|
family with sequence similarity 169, member B |
| chr10_+_19596214 | 2.74 |
ENSDART00000183110
|
CABZ01059626.1
|
|
| chr6_-_37749711 | 2.67 |
ENSDART00000078324
|
nipa1
|
non imprinted in Prader-Willi/Angelman syndrome 1 |
| chr3_-_32965848 | 2.66 |
ENSDART00000050930
|
casp6
|
caspase 6, apoptosis-related cysteine peptidase |
| chr20_-_23426339 | 2.62 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr1_+_12135129 | 2.57 |
ENSDART00000126020
|
spink4
|
serine peptidase inhibitor, Kazal type 4 |
| chr2_+_20539402 | 2.54 |
ENSDART00000129585
|
si:ch73-14h1.2
|
si:ch73-14h1.2 |
| chr21_-_43666420 | 2.54 |
ENSDART00000139008
ENSDART00000183996 ENSDART00000183395 |
si:dkey-229d11.3
si:dkey-229d11.5
|
si:dkey-229d11.3 si:dkey-229d11.5 |
| chr23_-_44574059 | 2.50 |
ENSDART00000123007
|
si:ch73-160p18.3
|
si:ch73-160p18.3 |
| chr14_+_34514336 | 2.48 |
ENSDART00000024440
|
foxi3b
|
forkhead box I3b |
| chr11_+_43431289 | 2.38 |
ENSDART00000192526
|
slc29a1b
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1b |
| chr20_-_33583779 | 2.35 |
ENSDART00000097823
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr16_-_17713859 | 2.30 |
ENSDART00000149275
|
zgc:174935
|
zgc:174935 |
| chr8_-_38022298 | 2.30 |
ENSDART00000067809
|
rab11fip1a
|
RAB11 family interacting protein 1 (class I) a |
| chr6_+_7466223 | 2.28 |
ENSDART00000148908
|
erbb3a
|
erb-b2 receptor tyrosine kinase 3a |
| chr11_-_25853212 | 2.18 |
ENSDART00000145655
|
tmem51b
|
transmembrane protein 51b |
| chr6_-_43449013 | 2.14 |
ENSDART00000122423
|
eevs
|
2-epi-5-epi-valiolone synthase |
| chr2_+_111919 | 2.11 |
ENSDART00000149391
|
fggy
|
FGGY carbohydrate kinase domain containing |
| chr4_-_77557279 | 2.08 |
ENSDART00000180113
|
AL935186.10
|
|
| chr14_+_48862987 | 2.05 |
ENSDART00000167810
|
zgc:154054
|
zgc:154054 |
| chr7_+_30178453 | 2.05 |
ENSDART00000174420
|
si:ch211-107o10.3
|
si:ch211-107o10.3 |
| chr17_+_3124129 | 2.00 |
ENSDART00000155323
|
zgc:136872
|
zgc:136872 |
| chr17_-_24575893 | 1.92 |
ENSDART00000141914
|
aftphb
|
aftiphilin b |
| chr12_+_48784731 | 1.90 |
ENSDART00000158348
|
zmiz1b
|
zinc finger, MIZ-type containing 1b |
| chr20_-_23439011 | 1.89 |
ENSDART00000022887
|
slc10a4
|
solute carrier family 10, member 4 |
| chr1_-_59313465 | 1.89 |
ENSDART00000158067
ENSDART00000159419 |
txndc11
|
thioredoxin domain containing 11 |
| chr19_-_18313303 | 1.87 |
ENSDART00000164644
ENSDART00000167480 ENSDART00000163104 |
si:dkey-208k4.2
|
si:dkey-208k4.2 |
| chr7_-_51773166 | 1.83 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr17_+_51906053 | 1.82 |
ENSDART00000159072
ENSDART00000056869 |
flvcr2a
|
feline leukemia virus subgroup C cellular receptor family, member 2a |
| chr10_+_9550419 | 1.80 |
ENSDART00000064977
|
si:ch211-243g18.2
|
si:ch211-243g18.2 |
| chr17_-_25331439 | 1.76 |
ENSDART00000155422
ENSDART00000082324 |
zpcx
|
zona pellucida protein C |
| chr3_+_18097700 | 1.71 |
ENSDART00000021634
|
wfikkn2a
|
info WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2a |
| chr3_-_55511569 | 1.71 |
ENSDART00000108995
|
tex2
|
testis expressed 2 |
| chr22_+_7480465 | 1.70 |
ENSDART00000034545
|
CELA1 (1 of many)
|
zgc:92745 |
| chr17_+_3128273 | 1.70 |
ENSDART00000122453
|
zgc:136872
|
zgc:136872 |
| chr3_+_48561112 | 1.67 |
ENSDART00000159682
|
slc16a5b
|
solute carrier family 16 (monocarboxylate transporter), member 5b |
| chr8_-_4892913 | 1.64 |
ENSDART00000126808
|
igsf9b
|
immunoglobulin superfamily, member 9b |
| chr4_-_68563862 | 1.63 |
ENSDART00000182970
|
BX548011.2
|
|
| chr7_+_67468999 | 1.61 |
ENSDART00000176368
|
rpl13
|
ribosomal protein L13 |
| chr5_-_57204352 | 1.57 |
ENSDART00000171252
ENSDART00000180727 |
man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chr12_-_990149 | 1.56 |
ENSDART00000054367
|
kdelr2b
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2b |
| chr14_-_26482096 | 1.55 |
ENSDART00000187280
|
smad5
|
SMAD family member 5 |
| chr8_+_15254564 | 1.53 |
ENSDART00000024433
|
slc5a9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr17_-_39886628 | 1.52 |
ENSDART00000002217
|
zmp:0000000545
|
zmp:0000000545 |
| chr12_-_7710563 | 1.52 |
ENSDART00000187382
|
CABZ01050062.1
|
|
| chr3_+_43086548 | 1.50 |
ENSDART00000163579
|
si:dkey-43p13.5
|
si:dkey-43p13.5 |
| chr2_-_58075414 | 1.48 |
ENSDART00000161920
|
nectin4
|
nectin cell adhesion molecule 4 |
| chr18_-_38271298 | 1.48 |
ENSDART00000143016
|
caprin1b
|
cell cycle associated protein 1b |
| chr25_-_16600811 | 1.43 |
ENSDART00000011397
|
cpa2
|
carboxypeptidase A2 (pancreatic) |
| chr14_+_26546175 | 1.42 |
ENSDART00000105932
|
si:dkeyp-110e4.11
|
si:dkeyp-110e4.11 |
| chr15_-_36365840 | 1.42 |
ENSDART00000192926
|
si:dkey-23k10.3
|
si:dkey-23k10.3 |
| chr9_-_33329700 | 1.38 |
ENSDART00000147265
ENSDART00000140039 |
rpl8
|
ribosomal protein L8 |
| chr23_+_26026383 | 1.35 |
ENSDART00000141553
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr5_+_6670945 | 1.33 |
ENSDART00000185686
|
pxna
|
paxillin a |
| chr14_+_3495542 | 1.32 |
ENSDART00000168934
|
gstp2
|
glutathione S-transferase pi 2 |
| chr15_-_19128705 | 1.32 |
ENSDART00000152428
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr21_+_39197628 | 1.30 |
ENSDART00000113607
|
cpdb
|
carboxypeptidase D, b |
| chr18_-_7539166 | 1.30 |
ENSDART00000133541
|
si:dkey-30c15.2
|
si:dkey-30c15.2 |
| chr4_+_77993977 | 1.29 |
ENSDART00000174118
|
terfa
|
telomeric repeat binding factor a |
| chr21_+_30351256 | 1.29 |
ENSDART00000078341
|
foxi3a
|
forkhead box I3a |
| chr21_+_39197418 | 1.27 |
ENSDART00000076000
|
cpdb
|
carboxypeptidase D, b |
| chr15_-_47929455 | 1.27 |
ENSDART00000064462
|
psma6l
|
proteasome subunit alpha 6, like |
| chr23_-_44677315 | 1.27 |
ENSDART00000143054
|
kif1c
|
kinesin family member 1C |
| chr22_-_22231720 | 1.26 |
ENSDART00000160165
|
ap3d1
|
adaptor-related protein complex 3, delta 1 subunit |
| chr20_+_30797329 | 1.25 |
ENSDART00000145066
|
nhsl1b
|
NHS-like 1b |
| chr16_+_41060161 | 1.25 |
ENSDART00000141130
|
scap
|
SREBF chaperone |
| chr11_-_45429199 | 1.25 |
ENSDART00000173111
|
rfc4
|
replication factor C (activator 1) 4 |
| chr1_-_59139599 | 1.24 |
ENSDART00000152233
|
si:ch1073-110a20.3
|
si:ch1073-110a20.3 |
| chr17_-_2596125 | 1.23 |
ENSDART00000175740
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr16_+_26547152 | 1.21 |
ENSDART00000141393
|
ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr15_-_43164591 | 1.20 |
ENSDART00000171305
|
ap1s3a
|
adaptor-related protein complex 1, sigma 3 subunit, a |
| chr3_+_49576662 | 1.18 |
ENSDART00000162275
|
CR394535.1
|
|
| chr22_-_15578402 | 1.18 |
ENSDART00000062986
|
hsh2d
|
hematopoietic SH2 domain containing |
| chr25_-_12730260 | 1.16 |
ENSDART00000171834
|
klhdc4
|
kelch domain containing 4 |
| chr2_-_722156 | 1.15 |
ENSDART00000045770
ENSDART00000169498 |
foxq1a
|
forkhead box Q1a |
| chr18_-_37241080 | 1.14 |
ENSDART00000126421
ENSDART00000078064 |
six9
|
SIX homeobox 9 |
| chr8_-_21184759 | 1.13 |
ENSDART00000139257
|
gls2a
|
glutaminase 2a (liver, mitochondrial) |
| chr3_+_49521106 | 1.12 |
ENSDART00000162799
|
crb3a
|
crumbs homolog 3a |
| chr6_+_7555614 | 1.11 |
ENSDART00000187590
|
myh10
|
myosin, heavy chain 10, non-muscle |
| chr23_+_2728095 | 1.11 |
ENSDART00000066086
|
zgc:114123
|
zgc:114123 |
| chr19_-_35439237 | 1.08 |
ENSDART00000145883
|
anln
|
anillin, actin binding protein |
| chr17_-_25330775 | 1.07 |
ENSDART00000154048
|
zpcx
|
zona pellucida protein C |
| chr3_+_32553714 | 1.06 |
ENSDART00000165638
|
pax10
|
paired box 10 |
| chr3_-_37758487 | 1.06 |
ENSDART00000150938
|
si:dkey-260c8.6
|
si:dkey-260c8.6 |
| chr16_+_27444098 | 1.04 |
ENSDART00000157690
|
invs
|
inversin |
| chr13_+_2894536 | 1.02 |
ENSDART00000183678
|
CABZ01087629.1
|
|
| chr10_-_3416258 | 1.01 |
ENSDART00000005168
|
ddx55
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 55 |
| chr21_-_20725853 | 1.00 |
ENSDART00000114502
|
si:ch211-22d5.2
|
si:ch211-22d5.2 |
| chr19_+_4061699 | 0.99 |
ENSDART00000158309
ENSDART00000166512 |
btr25
btr26
|
bloodthirsty-related gene family, member 25 bloodthirsty-related gene family, member 26 |
| chr25_+_5249513 | 0.99 |
ENSDART00000126814
|
CABZ01039863.1
|
|
| chr23_+_17387325 | 0.97 |
ENSDART00000083947
|
ptk6b
|
PTK6 protein tyrosine kinase 6b |
| chr1_-_24349759 | 0.97 |
ENSDART00000142740
ENSDART00000177989 |
lrba
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr10_+_4924388 | 0.96 |
ENSDART00000108595
|
slc46a2
|
solute carrier family 46 member 2 |
| chr8_-_4100365 | 0.96 |
ENSDART00000142846
|
cux2b
|
cut-like homeobox 2b |
| chr2_-_17694504 | 0.93 |
ENSDART00000133709
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr25_-_22889519 | 0.92 |
ENSDART00000128250
|
mob2a
|
MOB kinase activator 2a |
| chr7_-_26571994 | 0.91 |
ENSDART00000128801
|
si:dkey-62k3.6
|
si:dkey-62k3.6 |
| chr5_+_50913357 | 0.89 |
ENSDART00000092938
|
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr24_-_5911973 | 0.89 |
ENSDART00000077933
ENSDART00000077922 |
pimr64
|
Pim proto-oncogene, serine/threonine kinase, related 64 |
| chr5_-_68244564 | 0.89 |
ENSDART00000169350
|
CABZ01083944.1
|
|
| chr14_-_28052474 | 0.89 |
ENSDART00000172948
ENSDART00000135337 |
TSC22D3 (1 of many)
zgc:64189
|
si:ch211-220e11.3 zgc:64189 |
| chr19_+_31904836 | 0.88 |
ENSDART00000162297
ENSDART00000088340 ENSDART00000151280 ENSDART00000151218 |
tpd52
|
tumor protein D52 |
| chr22_-_38274188 | 0.87 |
ENSDART00000139420
ENSDART00000015117 |
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr3_-_41292569 | 0.87 |
ENSDART00000111856
|
sdk1a
|
sidekick cell adhesion molecule 1a |
| chr24_-_31904924 | 0.86 |
ENSDART00000156060
ENSDART00000129741 ENSDART00000154276 |
si:ch73-78o10.1
|
si:ch73-78o10.1 |
| chr8_+_25302172 | 0.85 |
ENSDART00000046182
ENSDART00000145316 |
gstm.3
|
glutathione S-transferase mu tandem duplicate 3 |
| chr19_+_4068134 | 0.85 |
ENSDART00000158285
|
btr26
|
bloodthirsty-related gene family, member 26 |
| chr18_-_7810214 | 0.84 |
ENSDART00000139505
ENSDART00000139188 |
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr16_+_40301056 | 0.83 |
ENSDART00000058578
|
rspo3
|
R-spondin 3 |
| chr21_-_217589 | 0.81 |
ENSDART00000185017
|
CZQB01146713.1
|
|
| chr23_-_45501177 | 0.81 |
ENSDART00000150103
|
col24a1
|
collagen type XXIV alpha 1 |
| chr6_+_56141852 | 0.78 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr22_-_209741 | 0.77 |
ENSDART00000171954
|
ora1
|
olfactory receptor class A related 1 |
| chr7_+_36484598 | 0.76 |
ENSDART00000173595
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr8_-_29658171 | 0.76 |
ENSDART00000147755
|
mpeg1.3
|
macrophage expressed 1, tandem duplicate 3 |
| chr2_-_57469115 | 0.75 |
ENSDART00000192201
|
pias4b
|
protein inhibitor of activated STAT, 4b |
| chr23_-_1144425 | 0.75 |
ENSDART00000098271
|
psmb11b
|
proteasome subunit beta11b |
| chr5_+_18014931 | 0.75 |
ENSDART00000142562
|
ascc2
|
activating signal cointegrator 1 complex subunit 2 |
| chr5_-_32505109 | 0.74 |
ENSDART00000188219
|
ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr6_-_55399214 | 0.74 |
ENSDART00000168367
|
ctsa
|
cathepsin A |
| chr17_+_26352372 | 0.74 |
ENSDART00000155177
|
grid1a
|
glutamate receptor, ionotropic, delta 1a |
| chr5_-_54712159 | 0.74 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr16_+_50953842 | 0.73 |
ENSDART00000174709
|
si:dkeyp-97a10.1
|
si:dkeyp-97a10.1 |
| chr10_+_37173029 | 0.73 |
ENSDART00000136510
|
ksr1a
|
kinase suppressor of ras 1a |
| chr17_+_1360192 | 0.73 |
ENSDART00000184561
|
siva1
|
SIVA1, apoptosis-inducing factor |
| chr23_+_27779452 | 0.72 |
ENSDART00000134785
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr16_+_2565433 | 0.72 |
ENSDART00000188014
ENSDART00000171378 |
preb
|
prolactin regulatory element binding |
| chr5_+_42259002 | 0.72 |
ENSDART00000083778
|
eral1
|
Era-like 12S mitochondrial rRNA chaperone 1 |
| chr16_-_2650341 | 0.71 |
ENSDART00000128169
ENSDART00000155432 ENSDART00000103722 |
lyplal1
|
lysophospholipase-like 1 |
| chr8_-_11170114 | 0.71 |
ENSDART00000133532
|
si:ch211-204d2.4
|
si:ch211-204d2.4 |
| chr12_-_2869565 | 0.71 |
ENSDART00000152772
|
zdhhc16b
|
zinc finger, DHHC-type containing 16b |
| chr13_-_51903150 | 0.71 |
ENSDART00000090644
|
mrpl2
|
mitochondrial ribosomal protein L2 |
| chr10_-_44467384 | 0.70 |
ENSDART00000186382
|
CABZ01072089.1
|
|
| chr7_-_45990681 | 0.70 |
ENSDART00000165441
|
si:ch211-260e23.7
|
si:ch211-260e23.7 |
| chr14_-_38865800 | 0.69 |
ENSDART00000173047
|
gsr
|
glutathione reductase |
| chr18_+_5308392 | 0.68 |
ENSDART00000179072
|
DUT
|
deoxyuridine triphosphatase |
| chr6_+_13506841 | 0.66 |
ENSDART00000032331
|
gmppab
|
GDP-mannose pyrophosphorylase Ab |
| chr2_-_17235891 | 0.66 |
ENSDART00000144251
|
artnb
|
artemin b |
| chr4_-_20483954 | 0.66 |
ENSDART00000187240
|
napepld
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr19_+_4800549 | 0.65 |
ENSDART00000056235
|
st3gal1l
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1, like |
| chr20_-_52902693 | 0.65 |
ENSDART00000166115
ENSDART00000161050 |
ctsbb
|
cathepsin Bb |
| chr20_-_41992878 | 0.65 |
ENSDART00000100967
|
si:dkeyp-114g9.1
|
si:dkeyp-114g9.1 |
| chr7_+_20917966 | 0.64 |
ENSDART00000129161
|
wrap53
|
WD repeat containing, antisense to TP53 |
| chr1_+_58290933 | 0.63 |
ENSDART00000140536
|
si:dkey-222h21.6
|
si:dkey-222h21.6 |
| chr1_-_43892349 | 0.63 |
ENSDART00000148416
|
tacr3a
|
tachykinin receptor 3a |
| chr1_+_40308077 | 0.61 |
ENSDART00000138992
|
vwa10.2
|
von Willebrand factor A domain containing 10, tandem duplicate 2 |
| chr10_+_1052591 | 0.61 |
ENSDART00000123405
|
unc5c
|
unc-5 netrin receptor C |
| chr15_-_434503 | 0.61 |
ENSDART00000122286
|
CABZ01056629.1
|
|
| chr16_-_31791165 | 0.60 |
ENSDART00000148389
|
chd4b
|
chromodomain helicase DNA binding protein 4b |
| chr7_+_38090515 | 0.59 |
ENSDART00000131387
|
cebpg
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr12_-_48312647 | 0.59 |
ENSDART00000114415
|
ascc1
|
activating signal cointegrator 1 complex subunit 1 |
| chr23_+_27778670 | 0.59 |
ENSDART00000053863
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr25_-_169291 | 0.58 |
ENSDART00000128344
|
lipcb
|
lipase, hepatic b |
| chr1_+_41609676 | 0.58 |
ENSDART00000183675
|
mogs
|
mannosyl-oligosaccharide glucosidase |
| chr17_-_52595932 | 0.58 |
ENSDART00000127225
|
si:ch211-173a9.7
|
si:ch211-173a9.7 |
| chr4_-_60312069 | 0.58 |
ENSDART00000167207
|
si:dkey-248e17.4
|
si:dkey-248e17.4 |
| chr20_-_25533739 | 0.58 |
ENSDART00000063064
|
cyp2ad6
|
cytochrome P450, family 2, subfamily AD, polypeptide 6 |
| chr12_-_30359031 | 0.57 |
ENSDART00000192628
|
tdrd1
|
tudor domain containing 1 |
| chr22_+_9091913 | 0.57 |
ENSDART00000186598
|
BX248395.1
|
|
| chr6_+_22326624 | 0.56 |
ENSDART00000020333
|
rae1
|
ribonucleic acid export 1 |
| chr7_+_24528430 | 0.56 |
ENSDART00000133022
|
si:dkeyp-75h12.2
|
si:dkeyp-75h12.2 |
| chr16_-_33001153 | 0.55 |
ENSDART00000147941
|
me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr11_-_4596431 | 0.55 |
ENSDART00000181615
|
FHIT
|
si:ch211-63i20.3 |
| chr2_+_43851983 | 0.55 |
ENSDART00000126413
|
nlrb5
|
NOD-like receptor family B, member 5 |
| chr23_+_25201077 | 0.54 |
ENSDART00000136675
|
si:dkey-151g10.3
|
si:dkey-151g10.3 |
| chr5_-_33959868 | 0.54 |
ENSDART00000143652
|
zgc:63972
|
zgc:63972 |
Network of associatons between targets according to the STRING database.
First level regulatory network of snai2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 10.8 | GO:0010873 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.9 | 2.8 | GO:0034380 | high-density lipoprotein particle assembly(GO:0034380) chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.9 | 3.8 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.9 | 5.1 | GO:0090134 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.8 | 4.0 | GO:0001773 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 0.5 | 1.8 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.5 | 1.8 | GO:0097037 | heme export(GO:0097037) |
| 0.4 | 2.4 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.4 | 2.9 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.4 | 5.8 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.4 | 2.9 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.3 | 1.3 | GO:0032208 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.3 | 4.6 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.3 | 1.1 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.3 | 1.6 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.3 | 1.0 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.3 | 4.6 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.3 | 4.3 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 0.7 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.2 | 1.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.2 | 3.3 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.2 | 3.0 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.2 | 10.4 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.2 | 0.8 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.2 | 1.1 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.2 | 0.7 | GO:0003400 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.2 | 0.7 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.2 | 1.3 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.2 | 2.7 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 0.5 | GO:1904088 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.2 | 0.5 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.4 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.1 | 1.3 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 1.1 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.1 | 1.1 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.1 | 1.9 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.8 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 0.7 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.7 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 1.5 | GO:2000223 | regulation of BMP signaling pathway involved in heart jogging(GO:2000223) |
| 0.1 | 0.6 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.1 | 1.9 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.4 | GO:1902746 | negative regulation of epithelial cell differentiation(GO:0030857) regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 0.8 | GO:0098815 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 1.1 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.1 | 13.0 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.1 | 2.3 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 1.2 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.1 | 1.4 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.9 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 5.8 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.1 | 2.9 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.1 | 0.6 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.1 | 0.4 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.1 | 0.7 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 3.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.1 | 0.6 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.1 | 1.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 1.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.4 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 0.5 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.5 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 0.5 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.1 | 0.4 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.1 | 0.4 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 2.9 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 0.2 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.1 | 0.3 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 0.3 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 1.3 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 0.6 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.5 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.1 | 0.2 | GO:0050787 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.1 | 0.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.4 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 1.7 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 1.1 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 0.2 | GO:0035046 | pronuclear migration(GO:0035046) |
| 0.1 | 12.6 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.0 | 7.4 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.5 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.5 | GO:0000022 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0034770 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.1 | GO:0042420 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.0 | 0.9 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 4.0 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 5.2 | GO:0045055 | regulated exocytosis(GO:0045055) |
| 0.0 | 0.6 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.5 | GO:1902307 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 1.0 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.3 | GO:0043217 | myelin maintenance(GO:0043217) |
| 0.0 | 0.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.4 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 4.7 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 2.6 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.9 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 1.9 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.8 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.3 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.3 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.3 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.7 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.5 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.3 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 1.0 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 2.3 | GO:0008544 | epidermis development(GO:0008544) |
| 0.0 | 0.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 1.1 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.9 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 1.9 | GO:0016485 | protein processing(GO:0016485) |
| 0.0 | 0.4 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.2 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.5 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.4 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.5 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.8 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 1.0 | 14.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.3 | 1.9 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.3 | 5.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.3 | 0.8 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.2 | 0.7 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.2 | 3.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 1.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.2 | 1.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.2 | 1.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 1.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 0.5 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 2.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 1.6 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.1 | 0.4 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 1.3 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 0.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 1.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.4 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.8 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 1.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.5 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 0.7 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 1.0 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 1.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 3.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 3.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.5 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 2.1 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.5 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 1.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 1.2 | GO:0030117 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.5 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 17.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.3 | GO:0005925 | focal adhesion(GO:0005925) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 10.8 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.9 | 2.8 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.5 | 1.6 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.5 | 4.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.5 | 3.3 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.5 | 2.3 | GO:0038131 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.4 | 5.8 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.4 | 2.9 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.4 | 2.9 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.4 | 5.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.4 | 4.1 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.3 | 3.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.3 | 1.9 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.3 | 2.3 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.3 | 1.8 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.3 | 0.8 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.3 | 1.5 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 0.7 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.2 | 2.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.2 | 1.8 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 0.6 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.2 | 22.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 0.7 | GO:0004475 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.2 | 1.3 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.2 | 10.4 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.2 | 4.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.2 | 0.5 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 4.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.6 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.7 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.1 | 1.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 2.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.8 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 3.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 4.6 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 0.5 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.1 | 1.6 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 0.6 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 1.5 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.4 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.1 | 0.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.7 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.7 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 2.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.6 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.7 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.8 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 1.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.2 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.0 | 0.5 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.0 | 0.4 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.5 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.5 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 2.1 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 1.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.9 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.6 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.4 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 2.1 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 1.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 5.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.4 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 1.3 | GO:0032934 | sterol binding(GO:0032934) |
| 0.0 | 1.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.5 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 1.8 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.0 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.7 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 1.0 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 2.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.3 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.4 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 1.0 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.3 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 1.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 1.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 1.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 3.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 2.7 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 1.7 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.3 | 2.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.3 | 5.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 4.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.2 | 2.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 3.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.2 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.1 | 0.7 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.8 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 1.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 4.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.7 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.7 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.6 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 3.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.5 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.1 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |