Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
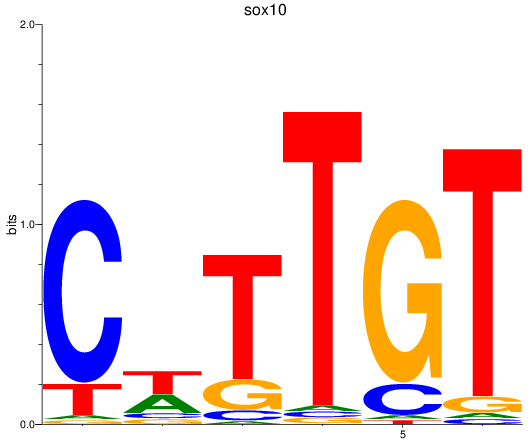
Results for sox10
Z-value: 1.04
Transcription factors associated with sox10
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox10
|
ENSDARG00000077467 | SRY-box transcription factor 10 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox10 | dr11_v1_chr3_+_1492174_1492174 | -0.74 | 1.1e-17 | Click! |
Activity profile of sox10 motif
Sorted Z-values of sox10 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_3629201 | 23.54 |
ENSDART00000136577
ENSDART00000132121 |
itih3a
|
inter-alpha-trypsin inhibitor heavy chain 3a |
| chr2_-_39017838 | 19.87 |
ENSDART00000048838
|
rbp2b
|
retinol binding protein 2b, cellular |
| chr1_-_19845378 | 15.38 |
ENSDART00000139314
ENSDART00000132958 ENSDART00000147502 |
grhprb
|
glyoxylate reductase/hydroxypyruvate reductase b |
| chr15_-_108414 | 13.74 |
ENSDART00000170044
|
apoa1b
|
apolipoprotein A-Ib |
| chr20_-_25518488 | 12.30 |
ENSDART00000186993
|
cyp2n13
|
cytochrome P450, family 2, subfamily N, polypeptide 13 |
| chr16_-_45917322 | 11.33 |
ENSDART00000060822
|
afp4
|
antifreeze protein type IV |
| chr22_-_23591493 | 10.73 |
ENSDART00000170266
|
f13b
|
coagulation factor XIII, B polypeptide |
| chr21_+_20771082 | 10.62 |
ENSDART00000079732
|
oxct1b
|
3-oxoacid CoA transferase 1b |
| chr5_+_28849155 | 10.54 |
ENSDART00000079090
|
zgc:174259
|
zgc:174259 |
| chr5_+_28848870 | 10.54 |
ENSDART00000149563
|
zgc:174259
|
zgc:174259 |
| chr8_-_50147948 | 10.19 |
ENSDART00000149010
|
hp
|
haptoglobin |
| chr22_-_23591340 | 10.09 |
ENSDART00000167024
|
f13b
|
coagulation factor XIII, B polypeptide |
| chr2_-_43653328 | 9.28 |
ENSDART00000037808
|
itgb1b.2
|
integrin, beta 1b.2 |
| chr21_-_25756119 | 9.21 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr15_-_32365940 | 8.81 |
ENSDART00000155371
|
c4
|
complement component 4 |
| chr23_+_17981127 | 8.67 |
ENSDART00000012571
ENSDART00000145200 |
chia.6
|
chitinase, acidic.6 |
| chr8_-_50482781 | 7.75 |
ENSDART00000056361
|
ido1
|
indoleamine 2,3-dioxygenase 1 |
| chr10_-_25823258 | 7.53 |
ENSDART00000064327
|
ftr54
|
finTRIM family, member 54 |
| chr3_-_18030938 | 7.42 |
ENSDART00000013540
|
si:ch73-141c7.1
|
si:ch73-141c7.1 |
| chr18_+_40462445 | 7.26 |
ENSDART00000087645
|
ugt5c2
|
UDP glucuronosyltransferase 5 family, polypeptide C2 |
| chr21_-_40083432 | 7.20 |
ENSDART00000141160
ENSDART00000191195 |
slc13a5a
|
info solute carrier family 13 (sodium-dependent citrate transporter), member 5a |
| chr2_-_20052561 | 7.17 |
ENSDART00000100133
|
dpydb
|
dihydropyrimidine dehydrogenase b |
| chr12_-_36045283 | 6.43 |
ENSDART00000160646
|
gprc5c
|
G protein-coupled receptor, class C, group 5, member C |
| chr23_+_10352921 | 6.19 |
ENSDART00000081193
|
KRT18 (1 of many)
|
si:ch211-133j6.3 |
| chr6_-_23931442 | 5.96 |
ENSDART00000160547
|
sec16b
|
SEC16 homolog B, endoplasmic reticulum export factor |
| chr8_-_38201415 | 5.88 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr24_-_2843107 | 5.60 |
ENSDART00000165290
|
cyb5a
|
cytochrome b5 type A (microsomal) |
| chr7_-_53117131 | 5.58 |
ENSDART00000169211
ENSDART00000168890 ENSDART00000172179 ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr16_+_26732086 | 5.47 |
ENSDART00000138496
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr3_+_15505275 | 5.42 |
ENSDART00000141714
|
nupr1
|
nuclear protein 1 |
| chr5_-_20123002 | 5.38 |
ENSDART00000026516
|
pxmp2
|
peroxisomal membrane protein 2 |
| chr9_+_8396755 | 5.37 |
ENSDART00000043067
|
zgc:171776
|
zgc:171776 |
| chr8_+_39802506 | 5.25 |
ENSDART00000018862
|
hnf1a
|
HNF1 homeobox a |
| chr1_-_33645967 | 4.92 |
ENSDART00000192758
|
cldng
|
claudin g |
| chr14_+_15155684 | 4.92 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr16_-_26731928 | 4.79 |
ENSDART00000135860
|
rnf41l
|
ring finger protein 41, like |
| chr4_-_77432218 | 4.79 |
ENSDART00000158683
|
slco1d1
|
solute carrier organic anion transporter family, member 1D1 |
| chr7_+_38255418 | 4.46 |
ENSDART00000052354
|
si:dkey-78a14.4
|
si:dkey-78a14.4 |
| chr6_+_30456788 | 4.42 |
ENSDART00000121492
|
FP236735.1
|
|
| chr4_+_25607743 | 4.41 |
ENSDART00000028297
|
acot14
|
acyl-CoA thioesterase 14 |
| chr7_+_49664174 | 4.40 |
ENSDART00000137059
ENSDART00000131210 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr16_-_41762983 | 4.38 |
ENSDART00000192936
|
si:dkey-199f5.8
|
si:dkey-199f5.8 |
| chr9_-_29497916 | 4.25 |
ENSDART00000060246
|
dnajc3a
|
DnaJ (Hsp40) homolog, subfamily C, member 3a |
| chr25_+_16880990 | 4.21 |
ENSDART00000020259
|
zgc:77158
|
zgc:77158 |
| chr16_+_19637384 | 4.19 |
ENSDART00000184773
ENSDART00000191895 ENSDART00000182020 ENSDART00000135359 |
macc1
|
metastasis associated in colon cancer 1 |
| chr19_-_27588842 | 4.19 |
ENSDART00000121643
|
si:dkeyp-46h3.2
|
si:dkeyp-46h3.2 |
| chr13_+_33651416 | 4.05 |
ENSDART00000180221
|
BX005372.1
|
|
| chr24_+_10027902 | 4.04 |
ENSDART00000175961
ENSDART00000172773 |
si:ch211-146l10.8
|
si:ch211-146l10.8 |
| chr20_-_43775495 | 4.03 |
ENSDART00000100610
ENSDART00000149001 ENSDART00000148809 ENSDART00000100608 |
matn3a
|
matrilin 3a |
| chr16_-_25607266 | 4.00 |
ENSDART00000192602
|
zgc:110410
|
zgc:110410 |
| chr16_+_40560622 | 3.93 |
ENSDART00000038294
|
tp53inp1
|
tumor protein p53 inducible nuclear protein 1 |
| chr7_-_71531846 | 3.90 |
ENSDART00000111797
|
acer2
|
alkaline ceramidase 2 |
| chr25_+_11281970 | 3.88 |
ENSDART00000180094
|
AKAP13
|
si:dkey-187e18.1 |
| chr3_-_16250527 | 3.79 |
ENSDART00000146699
ENSDART00000141181 |
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr5_+_44846434 | 3.69 |
ENSDART00000145299
ENSDART00000136521 |
kank1a
|
KN motif and ankyrin repeat domains 1a |
| chr14_+_46313135 | 3.62 |
ENSDART00000172902
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr6_-_39313027 | 3.61 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr15_-_34418525 | 3.58 |
ENSDART00000147582
|
agmo
|
alkylglycerol monooxygenase |
| chr17_-_25737452 | 3.55 |
ENSDART00000152021
|
si:ch211-214p16.3
|
si:ch211-214p16.3 |
| chr23_-_31506854 | 3.53 |
ENSDART00000131352
ENSDART00000138625 ENSDART00000133002 |
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr10_+_9550419 | 3.50 |
ENSDART00000064977
|
si:ch211-243g18.2
|
si:ch211-243g18.2 |
| chr1_+_12178215 | 3.49 |
ENSDART00000090380
|
stra6l
|
STRA6-like |
| chr22_+_1028724 | 3.47 |
ENSDART00000149625
|
si:ch73-352p18.4
|
si:ch73-352p18.4 |
| chr5_-_42894068 | 3.46 |
ENSDART00000169606
|
cxcl11.1
|
chemokine (C-X-C motif) ligand 11, duplicate 1 |
| chr10_+_16501699 | 3.42 |
ENSDART00000121864
|
slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr12_-_3237561 | 3.36 |
ENSDART00000164665
|
si:ch1073-13h15.3
|
si:ch1073-13h15.3 |
| chr11_-_10659195 | 3.34 |
ENSDART00000115255
|
mcf2l2
|
MCF.2 cell line derived transforming sequence-like 2 |
| chr15_-_36365840 | 3.32 |
ENSDART00000192926
|
si:dkey-23k10.3
|
si:dkey-23k10.3 |
| chr23_+_36063599 | 3.31 |
ENSDART00000103147
|
hoxc12a
|
homeobox C12a |
| chr23_+_36144487 | 3.28 |
ENSDART00000082473
|
hoxc3a
|
homeobox C3a |
| chr22_-_16180849 | 3.26 |
ENSDART00000090390
|
vcam1b
|
vascular cell adhesion molecule 1b |
| chr22_-_10541372 | 3.24 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr1_+_32051581 | 3.18 |
ENSDART00000146602
|
sts
|
steroid sulfatase (microsomal), isozyme S |
| chr18_-_14836862 | 3.14 |
ENSDART00000124843
|
mtss1la
|
metastasis suppressor 1-like a |
| chr15_-_17870090 | 3.12 |
ENSDART00000155066
|
atf5b
|
activating transcription factor 5b |
| chr7_-_39552314 | 3.09 |
ENSDART00000134174
|
slc22a18
|
solute carrier family 22, member 18 |
| chr10_-_40543896 | 3.09 |
ENSDART00000186921
ENSDART00000136031 |
taar18f
|
trace amine associated receptor 18f |
| chr10_+_7703251 | 3.07 |
ENSDART00000165134
|
ggcx
|
gamma-glutamyl carboxylase |
| chr17_+_24064014 | 3.01 |
ENSDART00000182782
ENSDART00000139063 ENSDART00000132755 |
b3gnt2b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2b |
| chr14_+_16036139 | 3.00 |
ENSDART00000190733
|
prelid1a
|
PRELI domain containing 1a |
| chr17_-_20236228 | 2.99 |
ENSDART00000136490
ENSDART00000029380 |
bnip4
|
BCL2 interacting protein 4 |
| chr14_-_33348221 | 2.99 |
ENSDART00000187749
|
rpl39
|
ribosomal protein L39 |
| chr3_+_17933553 | 2.99 |
ENSDART00000167731
ENSDART00000165644 |
cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr8_+_17869225 | 2.94 |
ENSDART00000080079
|
slc44a5b
|
solute carrier family 44, member 5b |
| chr20_+_25568694 | 2.93 |
ENSDART00000063107
ENSDART00000063128 |
cyp2p7
|
cytochrome P450, family 2, subfamily P, polypeptide 7 |
| chr15_+_22390076 | 2.93 |
ENSDART00000183764
|
oafa
|
OAF homolog a (Drosophila) |
| chr20_-_40360571 | 2.90 |
ENSDART00000144768
|
smpdl3a
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr19_+_31044487 | 2.89 |
ENSDART00000143494
|
ankmy2b
|
ankyrin repeat and MYND domain containing 2b |
| chr19_+_7636941 | 2.87 |
ENSDART00000081611
ENSDART00000163805 ENSDART00000112404 |
cgnb
|
cingulin b |
| chr22_-_38274188 | 2.86 |
ENSDART00000139420
ENSDART00000015117 |
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr9_+_15893093 | 2.83 |
ENSDART00000099483
ENSDART00000134657 |
si:dkey-14o1.20
|
si:dkey-14o1.20 |
| chr3_-_34069637 | 2.82 |
ENSDART00000151588
|
ighv9-1
|
immunoglobulin heavy variable 9-1 |
| chr11_+_12052791 | 2.78 |
ENSDART00000158479
|
si:ch211-156l18.8
|
si:ch211-156l18.8 |
| chr22_+_1911269 | 2.74 |
ENSDART00000164158
ENSDART00000168205 |
znf1156
|
zinc finger protein 1156 |
| chr1_+_38858399 | 2.71 |
ENSDART00000165454
|
CU915762.1
|
|
| chr5_+_26121393 | 2.71 |
ENSDART00000002221
|
bco2l
|
beta-carotene 15, 15-dioxygenase 2, like |
| chr15_+_31481939 | 2.70 |
ENSDART00000134306
|
or102-5
|
odorant receptor, family C, subfamily 102, member 5 |
| chr11_+_29965822 | 2.70 |
ENSDART00000127049
|
il1fma
|
interleukin-1 family member A |
| chr15_-_39955785 | 2.69 |
ENSDART00000154556
|
msh5
|
mutS homolog 5 |
| chr16_+_19029297 | 2.68 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr24_-_26369185 | 2.67 |
ENSDART00000080039
|
lrrc31
|
leucine rich repeat containing 31 |
| chr7_+_24814866 | 2.66 |
ENSDART00000173581
|
si:ch211-226l4.6
|
si:ch211-226l4.6 |
| chr20_-_54014539 | 2.65 |
ENSDART00000060466
|
si:dkey-241l7.6
|
si:dkey-241l7.6 |
| chr9_+_11293830 | 2.65 |
ENSDART00000144440
|
wnt6b
|
wingless-type MMTV integration site family, member 6b |
| chr14_-_24101897 | 2.62 |
ENSDART00000143695
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr12_-_42368296 | 2.61 |
ENSDART00000171075
|
zgc:111868
|
zgc:111868 |
| chr19_-_4785734 | 2.60 |
ENSDART00000113088
|
st3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr24_-_22003031 | 2.59 |
ENSDART00000133627
|
si:rp71-1f1.5
|
si:rp71-1f1.5 |
| chr25_+_35058088 | 2.57 |
ENSDART00000156838
|
zgc:112234
|
zgc:112234 |
| chr3_-_24458281 | 2.56 |
ENSDART00000153993
|
baiap2l2a
|
BAI1-associated protein 2-like 2a |
| chr17_-_6599484 | 2.54 |
ENSDART00000156927
|
ANKRD66
|
si:ch211-189e2.2 |
| chr17_+_30546579 | 2.54 |
ENSDART00000154385
|
nhsl1a
|
NHS-like 1a |
| chr16_+_29586468 | 2.50 |
ENSDART00000148926
|
mcl1b
|
MCL1, BCL2 family apoptosis regulator b |
| chr9_+_23003208 | 2.50 |
ENSDART00000021060
|
eaf2
|
ELL associated factor 2 |
| chr7_-_6459481 | 2.48 |
ENSDART00000173158
|
zgc:112234
|
zgc:112234 |
| chr7_+_61184104 | 2.48 |
ENSDART00000110671
|
zgc:194930
|
zgc:194930 |
| chr16_+_42465518 | 2.45 |
ENSDART00000058699
|
si:ch211-215k15.4
|
si:ch211-215k15.4 |
| chr22_+_25774750 | 2.45 |
ENSDART00000174421
|
AL929192.1
|
|
| chr3_-_9488350 | 2.45 |
ENSDART00000186707
|
FO904885.2
|
|
| chr2_+_19522082 | 2.45 |
ENSDART00000146098
|
pimr49
|
Pim proto-oncogene, serine/threonine kinase, related 49 |
| chr3_-_29977495 | 2.44 |
ENSDART00000077111
|
hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr18_-_40773413 | 2.44 |
ENSDART00000133797
|
vaspb
|
vasodilator stimulated phosphoprotein b |
| chr16_-_27566552 | 2.43 |
ENSDART00000142102
|
zgc:153215
|
zgc:153215 |
| chr11_-_11471857 | 2.39 |
ENSDART00000030103
|
krt94
|
keratin 94 |
| chr3_+_3545825 | 2.39 |
ENSDART00000109060
|
CR589947.1
|
|
| chr8_+_28467893 | 2.38 |
ENSDART00000189724
|
slc52a3
|
solute carrier family 52 (riboflavin transporter), member 3 |
| chr15_-_35410860 | 2.37 |
ENSDART00000191267
|
mecom
|
MDS1 and EVI1 complex locus |
| chr7_-_57332915 | 2.31 |
ENSDART00000162653
|
BX470176.1
|
|
| chr17_+_23994633 | 2.31 |
ENSDART00000156988
|
si:ch211-63b16.3
|
si:ch211-63b16.3 |
| chr3_+_3099492 | 2.31 |
ENSDART00000154156
|
si:dkey-30g5.1
|
si:dkey-30g5.1 |
| chr3_+_56645710 | 2.29 |
ENSDART00000193978
|
CR759836.1
|
|
| chr6_+_41039166 | 2.28 |
ENSDART00000125659
|
entpd8
|
ectonucleoside triphosphate diphosphohydrolase 8 |
| chr7_-_35126374 | 2.27 |
ENSDART00000141211
|
hsd11b2
|
hydroxysteroid (11-beta) dehydrogenase 2 |
| chr6_-_21091948 | 2.26 |
ENSDART00000057348
|
inha
|
inhibin, alpha |
| chr2_-_36604589 | 2.23 |
ENSDART00000167352
|
tradv41.0
|
T-cell receptor alpha/delta variable 41.0 |
| chr9_+_22359919 | 2.22 |
ENSDART00000009591
|
crygs4
|
crystallin, gamma S4 |
| chr2_-_17115256 | 2.21 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr19_-_9786914 | 2.20 |
ENSDART00000181669
|
si:dkey-14o18.2
|
si:dkey-14o18.2 |
| chr9_-_23891102 | 2.17 |
ENSDART00000186799
|
asb18
|
ankyrin repeat and SOCS box containing 18 |
| chr17_+_48164536 | 2.15 |
ENSDART00000161750
ENSDART00000156923 |
plekhd1
|
pleckstrin homology domain containing, family D (with coiled-coil domains) member 1 |
| chr9_-_18911608 | 2.14 |
ENSDART00000138785
|
si:dkey-239h2.3
|
si:dkey-239h2.3 |
| chr8_+_17868506 | 2.12 |
ENSDART00000175436
|
slc44a5b
|
solute carrier family 44, member 5b |
| chr8_+_2575993 | 2.12 |
ENSDART00000112914
|
si:ch211-51h9.7
|
si:ch211-51h9.7 |
| chr20_-_42534153 | 2.09 |
ENSDART00000061122
|
rfx6
|
regulatory factor X, 6 |
| chr10_-_35149513 | 2.08 |
ENSDART00000063434
ENSDART00000131291 |
ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr2_-_32768951 | 2.06 |
ENSDART00000004712
|
bfsp2
|
beaded filament structural protein 2, phakinin |
| chr21_-_25741411 | 2.05 |
ENSDART00000101211
|
cldnh
|
claudin h |
| chr18_-_22713621 | 2.04 |
ENSDART00000079013
|
si:ch73-113g13.2
|
si:ch73-113g13.2 |
| chr4_+_65607540 | 2.03 |
ENSDART00000192218
|
si:dkey-205i10.2
|
si:dkey-205i10.2 |
| chr2_+_19633493 | 2.02 |
ENSDART00000147989
|
pimr54
|
Pim proto-oncogene, serine/threonine kinase, related 54 |
| chr4_+_70414733 | 2.02 |
ENSDART00000162778
|
si:dkey-190j3.3
|
si:dkey-190j3.3 |
| chr4_+_17280868 | 2.02 |
ENSDART00000145349
|
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr23_-_1348933 | 1.99 |
ENSDART00000168981
|
CABZ01078120.1
|
|
| chr12_+_19030391 | 1.98 |
ENSDART00000153927
|
si:ch73-139e5.2
|
si:ch73-139e5.2 |
| chr15_+_43398317 | 1.96 |
ENSDART00000182528
ENSDART00000172154 ENSDART00000187688 |
actn4
|
actinin, alpha 4 |
| chr15_-_3736149 | 1.96 |
ENSDART00000182986
|
lpar6a
|
lysophosphatidic acid receptor 6a |
| chr24_-_26820698 | 1.96 |
ENSDART00000147788
|
fndc3bb
|
fibronectin type III domain containing 3Bb |
| chr7_+_27317174 | 1.96 |
ENSDART00000193058
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr5_+_26686639 | 1.94 |
ENSDART00000079064
|
tango2
|
transport and golgi organization 2 homolog (Drosophila) |
| chr5_+_40835601 | 1.94 |
ENSDART00000147767
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr25_-_27842654 | 1.93 |
ENSDART00000154852
ENSDART00000156906 |
asb15a
|
ankyrin repeat and SOCS box containing 15a |
| chr4_-_43731342 | 1.93 |
ENSDART00000146627
|
si:ch211-226o13.3
|
si:ch211-226o13.3 |
| chr4_+_25693463 | 1.93 |
ENSDART00000132864
|
acot18
|
acyl-CoA thioesterase 18 |
| chr14_-_9713549 | 1.93 |
ENSDART00000193356
ENSDART00000166739 |
si:zfos-2326c3.2
|
si:zfos-2326c3.2 |
| chr13_+_46941930 | 1.93 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr19_-_17208728 | 1.93 |
ENSDART00000151228
|
stmn1a
|
stathmin 1a |
| chr5_-_30380593 | 1.91 |
ENSDART00000148039
|
snx19a
|
sorting nexin 19a |
| chr18_+_33009828 | 1.90 |
ENSDART00000160100
|
olfcj1
|
olfactory receptor C family, j1 |
| chr1_-_59102320 | 1.90 |
ENSDART00000193379
|
si:zfos-2330d3.7
|
si:zfos-2330d3.7 |
| chr5_-_20205075 | 1.90 |
ENSDART00000051611
|
dao.3
|
D-amino-acid oxidase, tandem duplicate 3 |
| chr17_+_45305645 | 1.88 |
ENSDART00000172488
|
capn3a
|
calpain 3a, (p94) |
| chr11_+_20896122 | 1.87 |
ENSDART00000162339
ENSDART00000181111 |
CABZ01008730.1
|
|
| chr6_+_52891947 | 1.85 |
ENSDART00000174159
|
BX649282.3
|
|
| chr2_-_19576640 | 1.85 |
ENSDART00000141021
|
pimr51
|
Pim proto-oncogene, serine/threonine kinase, related 51 |
| chr14_+_46313396 | 1.84 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr7_+_10562118 | 1.83 |
ENSDART00000185188
ENSDART00000168801 |
zfand6
|
zinc finger, AN1-type domain 6 |
| chr21_-_2415808 | 1.82 |
ENSDART00000171179
|
si:ch211-241b2.5
|
si:ch211-241b2.5 |
| chr19_-_18152407 | 1.81 |
ENSDART00000193264
ENSDART00000016135 |
nfe2l3
|
nuclear factor, erythroid 2-like 3 |
| chr17_-_53440103 | 1.81 |
ENSDART00000156719
ENSDART00000156396 |
mycbp
|
c-myc binding protein |
| chr7_+_30240791 | 1.81 |
ENSDART00000109243
|
sema4bb
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Bb |
| chr5_+_57658898 | 1.80 |
ENSDART00000074268
ENSDART00000124568 |
zgc:153929
|
zgc:153929 |
| chr6_-_6254432 | 1.77 |
ENSDART00000081952
|
rtn4a
|
reticulon 4a |
| chr7_+_26058772 | 1.75 |
ENSDART00000101123
ENSDART00000173893 |
si:dkey-6n21.13
|
si:dkey-6n21.13 |
| chr5_-_54197084 | 1.75 |
ENSDART00000163640
|
grk1b
|
G protein-coupled receptor kinase 1 b |
| chr20_+_31217495 | 1.71 |
ENSDART00000020252
|
pdia6
|
protein disulfide isomerase family A, member 6 |
| chr23_-_29357764 | 1.71 |
ENSDART00000156512
|
si:ch211-129o18.4
|
si:ch211-129o18.4 |
| chr9_-_12811936 | 1.70 |
ENSDART00000188490
|
myo10l3
|
myosin X-like 3 |
| chr3_+_46764022 | 1.69 |
ENSDART00000023814
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr3_-_53114299 | 1.68 |
ENSDART00000109390
|
AL954361.1
|
|
| chr19_-_19721556 | 1.68 |
ENSDART00000165196
|
evx1
|
even-skipped homeobox 1 |
| chr7_-_24181159 | 1.68 |
ENSDART00000181206
|
zgc:153151
|
zgc:153151 |
| chr1_+_44173245 | 1.68 |
ENSDART00000159450
ENSDART00000106048 ENSDART00000157763 |
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr7_+_48555626 | 1.67 |
ENSDART00000125483
ENSDART00000083514 |
kcnq1
|
potassium voltage-gated channel, KQT-like subfamily, member 1 |
| chr3_-_29891218 | 1.67 |
ENSDART00000142118
|
slc25a39
|
solute carrier family 25, member 39 |
| chr5_+_69868911 | 1.67 |
ENSDART00000014649
ENSDART00000188215 ENSDART00000167385 |
ugt2a5
|
UDP glucuronosyltransferase 2 family, polypeptide A5 |
| chr21_-_40348790 | 1.66 |
ENSDART00000178123
|
CR847523.1
|
|
| chr21_-_41065369 | 1.66 |
ENSDART00000143749
|
larsb
|
leucyl-tRNA synthetase b |
| chr14_-_8787525 | 1.66 |
ENSDART00000160848
|
tgfb2l
|
transforming growth factor, beta 2, like |
| chr17_-_23609210 | 1.65 |
ENSDART00000064003
|
ifit15
|
interferon-induced protein with tetratricopeptide repeats 15 |
| chr7_+_22585447 | 1.65 |
ENSDART00000149144
|
chrnb1l
|
cholinergic receptor, nicotinic, beta 1 (muscle) like |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox10
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 13.7 | GO:0010872 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 2.4 | 7.2 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 2.1 | 10.6 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 1.2 | 6.0 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 1.0 | 4.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 1.0 | 7.7 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.9 | 5.6 | GO:0034334 | adherens junction maintenance(GO:0034334) mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.9 | 2.7 | GO:1902176 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 0.8 | 2.4 | GO:0072196 | proximal/distal pattern formation involved in nephron development(GO:0072047) proximal/distal pattern formation involved in pronephric nephron development(GO:0072196) |
| 0.8 | 8.7 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.7 | 3.6 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.7 | 4.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.7 | 3.5 | GO:0071938 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.7 | 2.1 | GO:0061178 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.7 | 6.5 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.6 | 4.1 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.5 | 3.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.5 | 2.0 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.5 | 1.5 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.5 | 1.9 | GO:0051352 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.5 | 1.9 | GO:0009078 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.4 | 2.4 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.4 | 3.5 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.4 | 1.1 | GO:2000638 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.4 | 1.8 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.4 | 11.3 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.4 | 1.4 | GO:0030149 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.3 | 17.1 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.3 | 2.3 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.3 | 6.1 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone metabolic process(GO:1901661) quinone biosynthetic process(GO:1901663) |
| 0.3 | 1.5 | GO:0035678 | neuromast hair cell morphogenesis(GO:0035678) |
| 0.3 | 9.3 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.3 | 2.9 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.3 | 2.9 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.3 | 1.4 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.3 | 1.7 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.3 | 3.9 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.3 | 1.4 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.3 | 1.6 | GO:1902946 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.3 | 1.4 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.3 | 2.9 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.3 | 5.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.3 | 1.6 | GO:0071405 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.2 | 2.7 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.2 | 20.8 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.2 | 0.9 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.2 | 0.9 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.2 | 2.5 | GO:2000403 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.2 | 1.0 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine metabolic process(GO:0046102) inosine biosynthetic process(GO:0046103) |
| 0.2 | 0.8 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.2 | 3.2 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.2 | 0.8 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.2 | 2.0 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.2 | 1.5 | GO:0006833 | water transport(GO:0006833) |
| 0.2 | 2.3 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.2 | 1.1 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.2 | 2.2 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.2 | 5.3 | GO:0002068 | glandular epithelial cell development(GO:0002068) type B pancreatic cell development(GO:0003323) |
| 0.2 | 2.5 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.2 | 0.5 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.2 | 16.4 | GO:0006956 | complement activation(GO:0006956) |
| 0.2 | 3.1 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.1 | 1.0 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 4.6 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.1 | 1.5 | GO:0032048 | cardiolipin metabolic process(GO:0032048) positive regulation of heart contraction(GO:0045823) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.1 | 1.8 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.1 | 0.6 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 1.4 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 6.1 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.1 | 1.2 | GO:0042664 | negative regulation of endodermal cell fate specification(GO:0042664) |
| 0.1 | 4.8 | GO:0071219 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.1 | 3.0 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.1 | 0.8 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.1 | 5.5 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.1 | 1.2 | GO:0099638 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.1 | 3.8 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.1 | 12.9 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 1.2 | GO:0051121 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 1.2 | GO:0030431 | sleep(GO:0030431) |
| 0.1 | 2.4 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.1 | 1.1 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.1 | 1.5 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 0.4 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 1.5 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 1.5 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 3.9 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 | 1.3 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 8.4 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.1 | 1.1 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.6 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.1 | 1.8 | GO:0072662 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 0.5 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 1.4 | GO:0098754 | detoxification(GO:0098754) |
| 0.1 | 1.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 1.5 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 1.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 3.3 | GO:0045104 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.1 | 1.1 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.1 | 4.3 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.1 | 1.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.8 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 0.8 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.1 | 6.3 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 1.5 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.1 | 1.7 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 0.6 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.1 | 0.8 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 2.0 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.3 | GO:0035093 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 6.9 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 2.6 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 1.7 | GO:0008589 | regulation of smoothened signaling pathway(GO:0008589) |
| 0.0 | 1.2 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 15.2 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 6.3 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.5 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 1.1 | GO:0035148 | tube formation(GO:0035148) |
| 0.0 | 0.2 | GO:0042311 | vasodilation(GO:0042311) |
| 0.0 | 0.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 2.7 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.3 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 1.1 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 3.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.7 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.4 | GO:1901642 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 1.5 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 2.1 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.0 | 1.1 | GO:0002548 | monocyte chemotaxis(GO:0002548) cellular response to interferon-gamma(GO:0071346) |
| 0.0 | 1.2 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.7 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.4 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 1.9 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.2 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 | 0.7 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.9 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 0.9 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 1.1 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.5 | GO:0006278 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.0 | 0.6 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.7 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.4 | GO:0019229 | regulation of vasoconstriction(GO:0019229) |
| 0.0 | 0.7 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.0 | 1.0 | GO:0031396 | regulation of protein ubiquitination(GO:0031396) |
| 0.0 | 0.4 | GO:0006101 | tricarboxylic acid cycle(GO:0006099) citrate metabolic process(GO:0006101) tricarboxylic acid metabolic process(GO:0072350) |
| 0.0 | 2.7 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.0 | 1.6 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 1.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.6 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 1.4 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 0.1 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 6.4 | GO:0070062 | extracellular exosome(GO:0070062) |
| 1.1 | 13.7 | GO:0042627 | chylomicron(GO:0042627) |
| 0.5 | 1.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.5 | 1.5 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.4 | 1.7 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.4 | 1.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.3 | 2.5 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.3 | 5.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.3 | 4.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 6.0 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.2 | 1.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 0.8 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.2 | 9.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.2 | 1.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.2 | 1.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 1.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.5 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.1 | 5.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 1.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 1.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 5.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.8 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 10.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 5.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 0.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 10.3 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 3.1 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 3.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 1.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 1.5 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.4 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 3.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 3.1 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.6 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 3.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 6.5 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 1.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 37.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 9.3 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.4 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 1.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 2.7 | GO:0000228 | nuclear chromosome(GO:0000228) |
| 0.0 | 0.5 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 4.8 | GO:0070161 | adherens junction(GO:0005912) anchoring junction(GO:0070161) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 13.6 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.9 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.0 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 9.1 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.5 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.1 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 15.4 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 2.7 | 13.7 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 2.6 | 7.7 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 2.1 | 10.6 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 1.9 | 9.3 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 1.6 | 6.5 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 1.4 | 7.2 | GO:0002061 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 1.3 | 5.4 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 1.2 | 7.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 1.0 | 3.0 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.9 | 5.5 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.8 | 7.4 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.8 | 8.7 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.8 | 3.1 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.5 | 4.2 | GO:0015154 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.5 | 2.0 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.5 | 3.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.5 | 1.5 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.5 | 1.4 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.5 | 1.9 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.4 | 2.2 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.4 | 5.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.4 | 3.0 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.4 | 1.3 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.4 | 2.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.4 | 3.6 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.4 | 2.4 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.4 | 1.4 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.3 | 1.7 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.3 | 3.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.3 | 2.7 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.3 | 1.5 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.3 | 5.9 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.3 | 2.6 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.3 | 3.9 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.3 | 1.7 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.3 | 1.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.3 | 6.3 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.3 | 17.1 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.3 | 1.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.2 | 2.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 2.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 4.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.2 | 0.9 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 1.4 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.2 | 3.8 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.2 | 5.9 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.2 | 0.8 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 0.8 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.2 | 3.8 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 1.7 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.2 | 2.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 3.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 1.4 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.2 | 0.8 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.2 | 1.0 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.2 | 1.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 0.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.2 | 1.2 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.2 | 20.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 1.5 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 12.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 4.4 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.2 | 8.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 2.7 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 0.4 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.1 | 3.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 2.0 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 1.7 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.5 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.1 | 1.0 | GO:0043394 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.1 | 1.4 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.8 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 1.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 7.5 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 6.0 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.1 | 0.4 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 6.5 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.1 | 2.3 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.1 | 1.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.9 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 1.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 1.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.5 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.1 | 1.1 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 1.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 3.2 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.1 | 0.8 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.9 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.2 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 1.5 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 1.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 1.1 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.1 | 0.8 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 1.6 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 2.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.3 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 4.0 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 1.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.6 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 11.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.0 | 1.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 3.9 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 5.6 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 4.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 21.2 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 1.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.8 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 1.1 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 1.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 4.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.4 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 1.2 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 1.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.2 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.0 | 1.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.5 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 11.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.3 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 1.6 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.7 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 1.4 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 1.1 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.1 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.7 | GO:0003774 | motor activity(GO:0003774) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.8 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 1.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 1.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 20.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 5.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 3.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 1.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 0.4 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 1.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 5.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 2.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.9 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 2.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.7 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 2.0 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.3 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 1.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 1.7 | 20.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.6 | 7.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.5 | 3.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.4 | 3.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.3 | 1.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.3 | 2.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.3 | 2.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.3 | 5.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 2.6 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 1.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 5.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 3.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 3.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 2.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 2.0 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 5.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 4.0 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 1.9 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 3.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 3.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 1.5 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.1 | 1.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 4.3 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 1.5 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.5 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.9 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |