Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
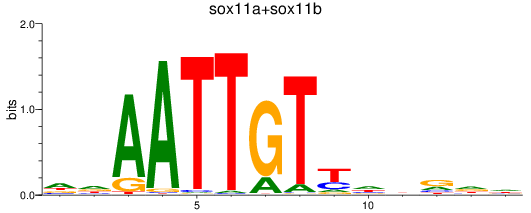
Results for sox11a+sox11b
Z-value: 0.74
Transcription factors associated with sox11a+sox11b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox11a
|
ENSDARG00000077811 | SRY-box transcription factor 11a |
|
sox11b
|
ENSDARG00000095743 | SRY-box transcription factor 11b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox11a | dr11_v1_chr17_-_35881841_35881841 | 0.52 | 6.9e-08 | Click! |
| sox11b | dr11_v1_chr20_-_30035326_30035326 | 0.34 | 8.5e-04 | Click! |
Activity profile of sox11a+sox11b motif
Sorted Z-values of sox11a+sox11b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_30282141 | 7.95 |
ENSDART00000122756
|
si:dkey-163f14.6
|
si:dkey-163f14.6 |
| chr21_-_41305748 | 7.27 |
ENSDART00000170457
|
nsg2
|
neuronal vesicle trafficking associated 2 |
| chr8_+_24861264 | 6.90 |
ENSDART00000099607
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr2_-_32643738 | 6.13 |
ENSDART00000112452
|
si:dkeyp-73d8.9
|
si:dkeyp-73d8.9 |
| chr17_-_48915427 | 5.62 |
ENSDART00000054781
|
lgals8b
|
galectin 8b |
| chr11_+_25596038 | 5.35 |
ENSDART00000140856
|
ccdc120
|
coiled-coil domain containing 120 |
| chr16_-_24598042 | 5.20 |
ENSDART00000156407
ENSDART00000154072 |
si:dkey-56f14.4
|
si:dkey-56f14.4 |
| chr2_+_16780643 | 5.20 |
ENSDART00000125647
ENSDART00000108611 ENSDART00000181245 ENSDART00000163194 |
tfa
|
transferrin-a |
| chr1_-_20928772 | 5.11 |
ENSDART00000078277
|
msmo1
|
methylsterol monooxygenase 1 |
| chr2_+_24177006 | 4.98 |
ENSDART00000132582
|
map4l
|
microtubule associated protein 4 like |
| chr16_-_22006996 | 4.97 |
ENSDART00000116114
|
si:dkey-71b5.7
|
si:dkey-71b5.7 |
| chr5_-_35301800 | 4.55 |
ENSDART00000085142
|
map1b
|
microtubule-associated protein 1B |
| chr19_+_26718074 | 4.52 |
ENSDART00000134455
|
zgc:100906
|
zgc:100906 |
| chr8_-_14050758 | 4.42 |
ENSDART00000133922
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr9_+_32978302 | 4.30 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| chr11_+_39672874 | 4.21 |
ENSDART00000046663
ENSDART00000157659 |
camta1b
|
calmodulin binding transcription activator 1b |
| chr10_+_21650828 | 4.09 |
ENSDART00000160754
|
pcdh1g1
|
protocadherin 1 gamma 1 |
| chr11_+_30513656 | 4.07 |
ENSDART00000008594
|
tmem178
|
transmembrane protein 178 |
| chr13_-_11644806 | 4.03 |
ENSDART00000169953
|
dctn1b
|
dynactin 1b |
| chr2_-_40135942 | 3.83 |
ENSDART00000176951
ENSDART00000098632 ENSDART00000148563 ENSDART00000149895 |
epha4a
|
eph receptor A4a |
| chr1_-_14234076 | 3.81 |
ENSDART00000040049
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr2_+_24177190 | 3.72 |
ENSDART00000099546
|
map4l
|
microtubule associated protein 4 like |
| chr9_+_17984358 | 3.48 |
ENSDART00000192399
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr5_+_66353750 | 3.41 |
ENSDART00000143410
|
si:ch211-261c8.5
|
si:ch211-261c8.5 |
| chr16_-_15988320 | 3.30 |
ENSDART00000160883
|
CABZ01060453.1
|
|
| chr13_-_40499296 | 3.24 |
ENSDART00000158338
|
CNNM1
|
Danio rerio cyclin and CBS domain divalent metal cation transport mediator 1 (cnnm1), mRNA. |
| chr3_-_18710009 | 3.19 |
ENSDART00000142478
|
grid2ipa
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, a |
| chr21_+_11401247 | 3.13 |
ENSDART00000143952
|
cel.1
|
carboxyl ester lipase, tandem duplicate 1 |
| chr13_+_13693722 | 3.11 |
ENSDART00000110509
|
si:ch211-194c3.5
|
si:ch211-194c3.5 |
| chr5_-_38342992 | 3.03 |
ENSDART00000140337
|
mink1
|
misshapen-like kinase 1 |
| chr4_+_14957360 | 3.03 |
ENSDART00000002770
ENSDART00000111882 ENSDART00000148292 |
tspan33a
|
tetraspanin 33a |
| chr19_-_21832441 | 3.02 |
ENSDART00000151272
ENSDART00000151442 ENSDART00000150168 ENSDART00000148797 ENSDART00000128196 ENSDART00000149259 ENSDART00000052556 ENSDART00000149658 ENSDART00000149639 ENSDART00000148424 |
mbpa
|
myelin basic protein a |
| chr4_+_28374628 | 3.01 |
ENSDART00000076037
|
alg10
|
asparagine-linked glycosylation 10 |
| chr15_-_20949692 | 2.93 |
ENSDART00000185548
|
tbcela
|
tubulin folding cofactor E-like a |
| chr12_+_26632448 | 2.86 |
ENSDART00000185762
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr1_+_16397063 | 2.83 |
ENSDART00000159794
|
micu3a
|
mitochondrial calcium uptake family, member 3a |
| chr5_+_26795465 | 2.82 |
ENSDART00000053001
|
tcn2
|
transcobalamin II |
| chr11_-_1409236 | 2.78 |
ENSDART00000121537
|
si:ch211-266k22.6
|
si:ch211-266k22.6 |
| chr6_+_4229360 | 2.77 |
ENSDART00000191347
ENSDART00000130642 |
FO082877.1
|
|
| chr16_+_36768674 | 2.71 |
ENSDART00000169208
ENSDART00000180470 |
si:ch73-215d9.1
|
si:ch73-215d9.1 |
| chr5_-_26118855 | 2.70 |
ENSDART00000009028
|
ela3l
|
elastase 3 like |
| chr20_-_26039841 | 2.69 |
ENSDART00000179929
|
si:dkey-12h9.6
|
si:dkey-12h9.6 |
| chr8_-_39978767 | 2.68 |
ENSDART00000083066
|
asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr1_+_10003193 | 2.68 |
ENSDART00000162675
|
trim2b
|
tripartite motif containing 2b |
| chr8_-_50888806 | 2.62 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr3_+_18807524 | 2.57 |
ENSDART00000055757
|
tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr17_-_43558494 | 2.50 |
ENSDART00000103830
|
nt5c1ab
|
5'-nucleotidase, cytosolic IAb |
| chr6_+_39812475 | 2.48 |
ENSDART00000067063
|
c1ql4b
|
complement component 1, q subcomponent-like 4b |
| chr1_+_26445615 | 2.46 |
ENSDART00000180810
|
g3bp2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr5_-_67911111 | 2.44 |
ENSDART00000051833
|
gsx1
|
GS homeobox 1 |
| chr23_+_40460333 | 2.44 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr23_-_17657348 | 2.43 |
ENSDART00000054736
|
bhlhe23
|
basic helix-loop-helix family, member e23 |
| chr19_-_31402429 | 2.42 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr19_-_42588510 | 2.39 |
ENSDART00000102583
|
sytl1
|
synaptotagmin-like 1 |
| chr18_-_26811959 | 2.39 |
ENSDART00000086250
|
znf592
|
zinc finger protein 592 |
| chr21_+_26389391 | 2.39 |
ENSDART00000077197
|
tmsb
|
thymosin, beta |
| chr20_-_47731768 | 2.27 |
ENSDART00000031167
|
tfap2d
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
| chr6_-_57539141 | 2.25 |
ENSDART00000156967
|
itcha
|
itchy E3 ubiquitin protein ligase a |
| chr10_-_24659822 | 2.21 |
ENSDART00000171842
ENSDART00000163102 |
proser1
|
proline and serine rich 1 |
| chr22_-_17606575 | 2.21 |
ENSDART00000183951
|
gpx4a
|
glutathione peroxidase 4a |
| chr4_-_16706776 | 2.18 |
ENSDART00000079461
|
dennd5b
|
DENN/MADD domain containing 5B |
| chr16_+_40217043 | 2.17 |
ENSDART00000191128
|
BX470182.1
|
|
| chr16_-_52540056 | 2.17 |
ENSDART00000188304
|
CR293507.1
|
|
| chr19_+_23919096 | 2.16 |
ENSDART00000090200
|
snapin
|
SNAP-associated protein |
| chr15_-_15357178 | 2.15 |
ENSDART00000106120
|
ywhag2
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 2 |
| chr22_+_3238474 | 2.13 |
ENSDART00000157954
|
si:ch1073-178p5.3
|
si:ch1073-178p5.3 |
| chr20_+_19793620 | 2.12 |
ENSDART00000136317
|
chrna2b
|
cholinergic receptor, nicotinic, alpha 2b (neuronal) |
| chr18_+_30028637 | 2.12 |
ENSDART00000139750
|
si:ch211-220f16.1
|
si:ch211-220f16.1 |
| chr2_-_1569250 | 2.06 |
ENSDART00000167202
|
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr15_+_16908085 | 2.04 |
ENSDART00000186870
|
ypel2b
|
yippee-like 2b |
| chr19_+_10592778 | 2.04 |
ENSDART00000135488
ENSDART00000151624 |
si:dkey-211g8.5
|
si:dkey-211g8.5 |
| chr9_-_29985390 | 2.03 |
ENSDART00000134157
|
il1rapl1a
|
interleukin 1 receptor accessory protein-like 1a |
| chr13_-_25198025 | 1.98 |
ENSDART00000159585
ENSDART00000144227 |
adka
|
adenosine kinase a |
| chr23_+_11669109 | 1.94 |
ENSDART00000091416
|
cntn3a.1
|
contactin 3a, tandem duplicate 1 |
| chr14_-_2187202 | 1.93 |
ENSDART00000160205
|
pcdh2ab12
|
protocadherin 2 alpha b 12 |
| chr12_-_18408566 | 1.92 |
ENSDART00000078767
ENSDART00000152748 |
tom1l2
|
target of myb1 like 2 membrane trafficking protein |
| chr7_-_40145097 | 1.92 |
ENSDART00000173634
|
wdr60
|
WD repeat domain 60 |
| chr17_+_33340675 | 1.91 |
ENSDART00000184396
ENSDART00000077553 |
xdh
|
xanthine dehydrogenase |
| chr1_+_26444986 | 1.90 |
ENSDART00000046376
|
g3bp2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr11_-_25733910 | 1.90 |
ENSDART00000171935
|
brpf3a
|
bromodomain and PHD finger containing, 3a |
| chr10_-_41907213 | 1.86 |
ENSDART00000167004
|
kdm2bb
|
lysine (K)-specific demethylase 2Bb |
| chr20_+_50852356 | 1.85 |
ENSDART00000167517
ENSDART00000168396 |
gphnb
|
gephyrin b |
| chr16_-_43233509 | 1.83 |
ENSDART00000025877
|
cldn12
|
claudin 12 |
| chr9_-_18568927 | 1.81 |
ENSDART00000084668
|
enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr9_+_29431763 | 1.79 |
ENSDART00000186095
ENSDART00000182640 |
uggt2
|
UDP-glucose glycoprotein glucosyltransferase 2 |
| chr5_-_57528943 | 1.72 |
ENSDART00000130320
|
pisd
|
phosphatidylserine decarboxylase |
| chr11_-_30158191 | 1.72 |
ENSDART00000155278
ENSDART00000156121 |
scml2
|
Scm polycomb group protein like 2 |
| chr10_-_7857494 | 1.70 |
ENSDART00000143215
|
inpp5ja
|
inositol polyphosphate-5-phosphatase Ja |
| chr2_+_26240631 | 1.66 |
ENSDART00000129895
|
palm1b
|
paralemmin 1b |
| chr25_-_29087925 | 1.66 |
ENSDART00000171758
|
rpp25a
|
ribonuclease P and MRP subunit p25, a |
| chr9_+_38883388 | 1.65 |
ENSDART00000135902
|
map2
|
microtubule-associated protein 2 |
| chr20_-_14718801 | 1.62 |
ENSDART00000137605
|
suco
|
SUN domain containing ossification factor |
| chr20_+_33519435 | 1.57 |
ENSDART00000061829
|
drc1
|
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
| chr5_-_2672492 | 1.56 |
ENSDART00000192337
|
CABZ01072548.1
|
|
| chr1_-_22512063 | 1.52 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr11_+_23760470 | 1.52 |
ENSDART00000175688
ENSDART00000121874 ENSDART00000086720 |
nfasca
|
neurofascin homolog (chicken) a |
| chr5_-_26795438 | 1.49 |
ENSDART00000146124
|
si:ch211-102c2.7
|
si:ch211-102c2.7 |
| chr18_-_16392229 | 1.49 |
ENSDART00000189031
|
mgat4c
|
mgat4 family, member C |
| chr25_-_20691609 | 1.47 |
ENSDART00000186942
|
edc3
|
enhancer of mRNA decapping 3 homolog (S. cerevisiae) |
| chr3_+_19299309 | 1.47 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr5_-_37871526 | 1.46 |
ENSDART00000136450
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr23_+_4226341 | 1.45 |
ENSDART00000012445
|
zgc:113278
|
zgc:113278 |
| chr2_+_22659787 | 1.45 |
ENSDART00000043956
|
zgc:161973
|
zgc:161973 |
| chr7_+_48297842 | 1.44 |
ENSDART00000052123
|
slc25a44b
|
solute carrier family 25, member 44 b |
| chr8_+_28358161 | 1.43 |
ENSDART00000062682
|
adipor1b
|
adiponectin receptor 1b |
| chr5_-_24869213 | 1.42 |
ENSDART00000112287
ENSDART00000144635 |
gas2l1
|
growth arrest-specific 2 like 1 |
| chr7_+_54642005 | 1.40 |
ENSDART00000171864
|
fgf19
|
fibroblast growth factor 19 |
| chr15_-_40267485 | 1.39 |
ENSDART00000152253
|
kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr11_+_16153207 | 1.37 |
ENSDART00000192356
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr11_-_25734417 | 1.35 |
ENSDART00000103570
|
brpf3a
|
bromodomain and PHD finger containing, 3a |
| chr12_+_14676349 | 1.33 |
ENSDART00000143401
|
becn1
|
beclin 1, autophagy related |
| chr23_-_24234371 | 1.31 |
ENSDART00000124539
|
ddost
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) |
| chr8_-_1838315 | 1.31 |
ENSDART00000114476
ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr20_+_44311448 | 1.30 |
ENSDART00000114660
|
opn8b
|
opsin 8, group member b |
| chr19_-_9522548 | 1.28 |
ENSDART00000045245
|
ing4
|
inhibitor of growth family, member 4 |
| chr23_-_25126003 | 1.26 |
ENSDART00000034953
|
idh3g
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr5_+_61657010 | 1.24 |
ENSDART00000050916
|
ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr22_+_7439186 | 1.23 |
ENSDART00000190667
|
zgc:92041
|
zgc:92041 |
| chr10_+_42521943 | 1.22 |
ENSDART00000010420
ENSDART00000075303 |
actr1
|
ARP1 actin related protein 1, centractin |
| chr22_-_6219674 | 1.19 |
ENSDART00000182582
|
si:ch211-274k16.2
|
si:ch211-274k16.2 |
| chr5_+_63857055 | 1.17 |
ENSDART00000138950
|
rgs3b
|
regulator of G protein signaling 3b |
| chrM_+_12897 | 1.17 |
ENSDART00000093622
|
mt-nd5
|
NADH dehydrogenase 5, mitochondrial |
| chr11_-_16093018 | 1.13 |
ENSDART00000139309
ENSDART00000139819 |
SPATA1
|
si:dkey-205k8.5 |
| chr9_+_29520696 | 1.13 |
ENSDART00000144430
|
fdx1
|
ferredoxin 1 |
| chr11_+_41243094 | 1.11 |
ENSDART00000191782
ENSDART00000169817 ENSDART00000162157 |
pax7a
|
paired box 7a |
| chr5_+_24543862 | 1.08 |
ENSDART00000029699
|
atp6v0a2b
|
ATPase H+ transporting V0 subunit a2b |
| chr20_+_54383838 | 1.07 |
ENSDART00000157737
|
lrfn5b
|
leucine rich repeat and fibronectin type III domain containing 5b |
| chr24_-_38657683 | 1.07 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr10_-_28761454 | 1.04 |
ENSDART00000129400
|
alcama
|
activated leukocyte cell adhesion molecule a |
| chr22_+_7462997 | 0.96 |
ENSDART00000106082
|
zgc:112368
|
zgc:112368 |
| chr19_-_18135724 | 0.96 |
ENSDART00000186609
|
cbx3a
|
chromobox homolog 3a (HP1 gamma homolog, Drosophila) |
| chr19_-_6083761 | 0.96 |
ENSDART00000151185
ENSDART00000143941 |
gsk3aa
|
glycogen synthase kinase 3 alpha a |
| chr6_-_27108844 | 0.94 |
ENSDART00000073883
|
dtymk
|
deoxythymidylate kinase (thymidylate kinase) |
| chr21_-_21530868 | 0.92 |
ENSDART00000174231
|
or133-9
|
odorant receptor, family H, subfamily 133, member 9 |
| chr6_-_1768724 | 0.89 |
ENSDART00000162488
ENSDART00000163613 |
zgc:158417
|
zgc:158417 |
| chr15_-_14467394 | 0.88 |
ENSDART00000191944
|
numbl
|
numb homolog (Drosophila)-like |
| chr19_-_17864213 | 0.88 |
ENSDART00000151043
|
ints8
|
integrator complex subunit 8 |
| chr9_+_32859967 | 0.88 |
ENSDART00000168992
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr5_-_24543526 | 0.87 |
ENSDART00000046384
|
trmt2a
|
tRNA methyltransferase 2 homolog A |
| chr20_+_50115335 | 0.86 |
ENSDART00000031139
|
slc24a4b
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4b |
| chr21_-_18648861 | 0.86 |
ENSDART00000112113
|
si:dkey-112m2.1
|
si:dkey-112m2.1 |
| chr12_-_15567104 | 0.85 |
ENSDART00000053003
|
hexim1
|
hexamethylene bis-acetamide inducible 1 |
| chr10_+_17714866 | 0.85 |
ENSDART00000039969
|
slc20a1b
|
solute carrier family 20 (phosphate transporter), member 1b |
| chr11_+_13024002 | 0.84 |
ENSDART00000104113
|
btf3l4
|
basic transcription factor 3-like 4 |
| chr8_-_45867358 | 0.84 |
ENSDART00000132810
|
adam9
|
ADAM metallopeptidase domain 9 |
| chr20_+_41664350 | 0.82 |
ENSDART00000186247
|
fam184a
|
family with sequence similarity 184, member A |
| chr6_-_15492030 | 0.82 |
ENSDART00000156141
ENSDART00000183992 |
st6gal2b
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2b |
| chr5_+_3118231 | 0.81 |
ENSDART00000179809
|
supt4h1
|
SPT4 homolog, DSIF elongation factor subunit |
| chr25_-_7764083 | 0.81 |
ENSDART00000179800
ENSDART00000181858 |
phf21ab
|
PHD finger protein 21Ab |
| chr21_+_42930558 | 0.80 |
ENSDART00000135234
|
stk32a
|
serine/threonine kinase 32A |
| chr5_+_41996889 | 0.80 |
ENSDART00000097580
|
pigl
|
phosphatidylinositol glycan anchor biosynthesis, class L |
| chr10_+_38806719 | 0.79 |
ENSDART00000184416
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr10_-_27009413 | 0.79 |
ENSDART00000139942
ENSDART00000146983 ENSDART00000132352 |
uqcc3
|
ubiquinol-cytochrome c reductase complex assembly factor 3 |
| chr22_-_10440688 | 0.76 |
ENSDART00000111962
|
nol8
|
nucleolar protein 8 |
| chr5_-_13251907 | 0.75 |
ENSDART00000176774
ENSDART00000030553 |
top3b
|
DNA topoisomerase III beta |
| chr2_+_9061885 | 0.72 |
ENSDART00000028906
|
pigk
|
phosphatidylinositol glycan anchor biosynthesis, class K |
| chr6_-_31336317 | 0.72 |
ENSDART00000193927
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr21_+_26612777 | 0.71 |
ENSDART00000142667
|
esrra
|
estrogen-related receptor alpha |
| chr4_-_25859305 | 0.70 |
ENSDART00000159528
|
nr2c1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr22_+_6674992 | 0.68 |
ENSDART00000144054
|
si:ch211-209l18.2
|
si:ch211-209l18.2 |
| chr14_-_17576662 | 0.68 |
ENSDART00000193893
|
rnf4
|
ring finger protein 4 |
| chr8_-_39822917 | 0.67 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr10_+_17713022 | 0.62 |
ENSDART00000170453
|
slc20a1b
|
solute carrier family 20 (phosphate transporter), member 1b |
| chr2_+_47906240 | 0.61 |
ENSDART00000122206
|
ftr23
|
finTRIM family, member 23 |
| chr22_+_10440991 | 0.60 |
ENSDART00000064805
|
cenpp
|
centromere protein P |
| chr17_-_43031763 | 0.60 |
ENSDART00000132754
ENSDART00000050399 |
npc2
|
Niemann-Pick disease, type C2 |
| chr2_-_30055432 | 0.60 |
ENSDART00000056747
|
shhb
|
sonic hedgehog b |
| chr21_-_40317035 | 0.59 |
ENSDART00000143648
ENSDART00000013359 |
or101-1
|
odorant receptor, family B, subfamily 101, member 1 |
| chr8_+_13368150 | 0.56 |
ENSDART00000114699
|
slc5a5
|
solute carrier family 5 (sodium/iodide cotransporter), member 5 |
| chr18_+_3579829 | 0.52 |
ENSDART00000158763
ENSDART00000182850 ENSDART00000162754 ENSDART00000178789 ENSDART00000172656 |
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr17_+_12658411 | 0.52 |
ENSDART00000139918
|
gpn1
|
GPN-loop GTPase 1 |
| chr25_-_20378721 | 0.52 |
ENSDART00000181707
|
kctd15a
|
potassium channel tetramerization domain containing 15a |
| chr25_+_19485198 | 0.51 |
ENSDART00000156730
|
glsl
|
glutaminase like |
| chr23_-_31403668 | 0.50 |
ENSDART00000147498
|
CR759830.1
|
|
| chr1_-_23596391 | 0.47 |
ENSDART00000155184
|
lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr12_+_32323098 | 0.47 |
ENSDART00000188722
|
ANKFN1
|
si:ch211-277e21.2 |
| chr19_+_2279051 | 0.46 |
ENSDART00000182103
|
itgb8
|
integrin, beta 8 |
| chr10_+_2587234 | 0.46 |
ENSDART00000126937
|
wu:fb59d01
|
wu:fb59d01 |
| chr1_+_21309386 | 0.45 |
ENSDART00000054442
ENSDART00000145661 |
med28
|
mediator complex subunit 28 |
| chr10_+_24660017 | 0.45 |
ENSDART00000079597
|
vps36
|
vacuolar protein sorting 36 homolog (S. cerevisiae) |
| chr7_+_19762595 | 0.44 |
ENSDART00000130347
|
si:dkey-9k7.3
|
si:dkey-9k7.3 |
| chr4_-_18635005 | 0.43 |
ENSDART00000125361
|
pparaa
|
peroxisome proliferator-activated receptor alpha a |
| chr4_-_52165969 | 0.42 |
ENSDART00000171130
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr2_-_37532772 | 0.42 |
ENSDART00000133951
|
arhgef18a
|
rho/rac guanine nucleotide exchange factor (GEF) 18a |
| chr11_-_891674 | 0.42 |
ENSDART00000172904
|
atg7
|
ATG7 autophagy related 7 homolog (S. cerevisiae) |
| chr7_+_56735195 | 0.41 |
ENSDART00000082830
|
KIAA0895L
|
KIAA0895 like |
| chr10_-_39321367 | 0.41 |
ENSDART00000129647
|
smtlb
|
somatolactin beta |
| chr17_-_27223965 | 0.41 |
ENSDART00000192577
|
asap3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| chr8_+_34434345 | 0.39 |
ENSDART00000190246
ENSDART00000189447 ENSDART00000185557 ENSDART00000189230 |
zgc:174461
|
zgc:174461 |
| chr19_-_47456787 | 0.38 |
ENSDART00000168792
|
tfap2e
|
transcription factor AP-2 epsilon |
| chr17_+_47090497 | 0.35 |
ENSDART00000169038
ENSDART00000159292 |
zgc:103755
|
zgc:103755 |
| chr15_-_45510977 | 0.35 |
ENSDART00000090596
|
fgf12b
|
fibroblast growth factor 12b |
| chr20_+_9128829 | 0.34 |
ENSDART00000064144
ENSDART00000137450 |
bpnt1
|
bisphosphate nucleotidase 1 |
| chr10_-_41906609 | 0.34 |
ENSDART00000102530
|
kdm2bb
|
lysine (K)-specific demethylase 2Bb |
| chr7_+_57108823 | 0.33 |
ENSDART00000184943
ENSDART00000055956 |
enosf1
|
enolase superfamily member 1 |
| chr4_-_59057046 | 0.33 |
ENSDART00000150249
|
znf1132
|
zinc finger protein 1132 |
| chr3_-_32873641 | 0.33 |
ENSDART00000075277
|
zgc:113090
|
zgc:113090 |
| chr14_+_35383267 | 0.32 |
ENSDART00000143818
|
clint1a
|
clathrin interactor 1a |
| chr22_-_35961365 | 0.31 |
ENSDART00000188434
|
LO016987.2
|
|
| chr19_+_32401278 | 0.28 |
ENSDART00000184353
|
atxn1a
|
ataxin 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox11a+sox11b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.9 | GO:0015824 | proline transport(GO:0015824) |
| 1.4 | 4.1 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.9 | 2.8 | GO:0006824 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.7 | 7.6 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.6 | 1.9 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.6 | 1.9 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.5 | 3.8 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.5 | 2.7 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.4 | 1.3 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.4 | 8.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.4 | 1.8 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.3 | 1.4 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.3 | 2.1 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.3 | 2.0 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.3 | 4.5 | GO:0001840 | neural plate development(GO:0001840) |
| 0.3 | 0.9 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.3 | 3.0 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.3 | 3.1 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.3 | 1.1 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.3 | 2.9 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.3 | 4.5 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.2 | 1.5 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.2 | 1.9 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.2 | 3.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.2 | 7.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.2 | 1.1 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.2 | 1.7 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.2 | 1.6 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.2 | 1.6 | GO:0003352 | regulation of cilium movement(GO:0003352) |
| 0.2 | 2.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.2 | 1.7 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.2 | 2.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.2 | 2.1 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.2 | 3.1 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.2 | 2.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.2 | 1.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.2 | 2.8 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 | 4.0 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 1.5 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 1.4 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.7 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 1.1 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.1 | 0.7 | GO:0097065 | anterior head development(GO:0097065) |
| 0.1 | 0.8 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 2.0 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.1 | 0.8 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 0.7 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 5.1 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.1 | 2.6 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 0.6 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 2.7 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 3.0 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.1 | 0.3 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.1 | 1.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 1.1 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 0.4 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 2.3 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 0.5 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.1 | 1.5 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 1.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 0.4 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 | 1.9 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 0.5 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.4 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.4 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 1.4 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.8 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 1.0 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 3.3 | GO:0034504 | protein localization to nucleus(GO:0034504) |
| 0.0 | 0.8 | GO:0060324 | face development(GO:0060324) |
| 0.0 | 1.0 | GO:0097192 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.0 | 0.9 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.8 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 1.0 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.9 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 1.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 6.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.3 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 1.7 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.3 | GO:0045682 | regulation of epidermis development(GO:0045682) |
| 0.0 | 1.5 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 1.3 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 2.4 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 3.0 | GO:0051604 | protein maturation(GO:0051604) |
| 0.0 | 0.7 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.0 | 8.8 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
| 0.0 | 0.2 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.0 | 1.5 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 1.5 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 4.1 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 2.7 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.1 | GO:0042304 | regulation of fatty acid biosynthetic process(GO:0042304) |
| 0.0 | 1.4 | GO:0008360 | regulation of cell shape(GO:0008360) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.3 | 1.3 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.2 | 1.7 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 2.8 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 3.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.2 | 4.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 3.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.2 | 1.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 1.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.7 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 2.7 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 1.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 2.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.5 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 1.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.8 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 1.6 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.1 | 0.3 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 5.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 16.3 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 1.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 1.7 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 2.4 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 3.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.2 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 3.7 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 5.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.9 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 4.4 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 7.4 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 16.3 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.5 | GO:0070382 | exocytic vesicle(GO:0070382) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.7 | 5.1 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.7 | 2.0 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.6 | 1.9 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.5 | 7.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.5 | 1.5 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.4 | 2.2 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.4 | 2.0 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.4 | 1.5 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.4 | 1.8 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.4 | 2.8 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.3 | 0.9 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.3 | 0.8 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.3 | 1.3 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.2 | 2.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 2.6 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.2 | 3.2 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.2 | 3.8 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 1.7 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.2 | 1.9 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.2 | 3.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 1.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.2 | 4.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 2.6 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 1.8 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 0.4 | GO:0005131 | growth hormone receptor binding(GO:0005131) growth hormone activity(GO:0070186) |
| 0.1 | 4.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 3.0 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 2.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 2.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.8 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 1.5 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 0.9 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 1.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.4 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 2.7 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 3.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.2 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.1 | 1.1 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.6 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.6 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 0.8 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 1.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 17.8 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 1.5 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 1.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 4.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 7.4 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 1.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 1.1 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 1.4 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 5.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 2.1 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 0.9 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 1.5 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.6 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.9 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 4.9 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.6 | GO:0015485 | cholesterol binding(GO:0015485) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.9 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.9 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 5.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 1.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 1.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 1.4 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 1.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 1.8 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 1.6 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 3.0 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 1.7 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 1.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 2.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 0.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.8 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 4.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |