Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
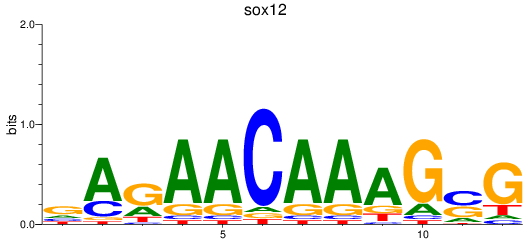
Results for sox12
Z-value: 0.96
Transcription factors associated with sox12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox12
|
ENSDARG00000025847 | SRY-box transcription factor 12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox12 | dr11_v1_chr11_-_24191928_24191928 | 0.86 | 2.3e-28 | Click! |
Activity profile of sox12 motif
Sorted Z-values of sox12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_44280061 | 13.70 |
ENSDART00000136818
|
mpz
|
myelin protein zero |
| chr20_-_39265453 | 11.72 |
ENSDART00000141193
|
clu
|
clusterin |
| chr22_-_37349967 | 11.21 |
ENSDART00000104493
|
sox2
|
SRY (sex determining region Y)-box 2 |
| chr16_-_9897613 | 11.08 |
ENSDART00000104058
|
ncalda
|
neurocalcin delta a |
| chr4_+_21129752 | 9.29 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr19_-_19339285 | 9.24 |
ENSDART00000158413
ENSDART00000170479 |
cspg5b
|
chondroitin sulfate proteoglycan 5b |
| chr2_+_24188502 | 7.84 |
ENSDART00000181955
ENSDART00000191706 ENSDART00000125909 |
map4l
|
microtubule associated protein 4 like |
| chr9_-_31278048 | 7.62 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr6_+_36942966 | 7.18 |
ENSDART00000028895
|
negr1
|
neuronal growth regulator 1 |
| chr4_-_2525916 | 7.13 |
ENSDART00000134123
ENSDART00000132581 ENSDART00000019508 |
csrp2
|
cysteine and glycine-rich protein 2 |
| chr19_+_8481802 | 7.07 |
ENSDART00000050217
|
efna1a
|
ephrin-A1a |
| chr5_-_38384755 | 7.05 |
ENSDART00000188573
ENSDART00000051233 |
mink1
|
misshapen-like kinase 1 |
| chr20_+_30445971 | 6.84 |
ENSDART00000153150
|
myt1la
|
myelin transcription factor 1-like, a |
| chr12_+_5081759 | 6.27 |
ENSDART00000164178
|
prrt2
|
proline-rich transmembrane protein 2 |
| chr10_+_26834985 | 6.24 |
ENSDART00000147518
|
fam89b
|
family with sequence similarity 89, member B |
| chr22_+_21618121 | 6.17 |
ENSDART00000133939
|
tle2a
|
transducin like enhancer of split 2a |
| chr6_+_27667359 | 6.11 |
ENSDART00000159624
ENSDART00000049177 |
rab6ba
|
RAB6B, member RAS oncogene family a |
| chr8_+_26859639 | 6.10 |
ENSDART00000133440
|
prdm2a
|
PR domain containing 2, with ZNF domain a |
| chr7_-_38861741 | 6.03 |
ENSDART00000173629
ENSDART00000037361 ENSDART00000173953 |
phf21aa
|
PHD finger protein 21Aa |
| chr21_+_31150438 | 5.70 |
ENSDART00000065366
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr16_-_9897769 | 5.63 |
ENSDART00000187044
|
ncalda
|
neurocalcin delta a |
| chr7_+_22680560 | 5.58 |
ENSDART00000133761
|
ponzr4
|
plac8 onzin related protein 4 |
| chr2_-_24348948 | 5.45 |
ENSDART00000136559
|
ano8a
|
anoctamin 8a |
| chr14_-_33297287 | 5.37 |
ENSDART00000045555
ENSDART00000138294 ENSDART00000075056 |
rab41
|
RAB41, member RAS oncogene family |
| chr20_-_2355357 | 5.16 |
ENSDART00000085281
|
EPB41L2
|
si:ch73-18b11.1 |
| chr2_-_24348642 | 5.07 |
ENSDART00000181739
|
ano8a
|
anoctamin 8a |
| chr5_+_26221011 | 5.06 |
ENSDART00000007587
|
gtf2h2
|
general transcription factor IIH, polypeptide 2 |
| chr8_+_42998944 | 4.94 |
ENSDART00000048819
|
rassf2a
|
Ras association (RalGDS/AF-6) domain family member 2a |
| chr17_+_37624704 | 4.82 |
ENSDART00000157001
|
tmem229b
|
transmembrane protein 229B |
| chr14_-_1955257 | 4.51 |
ENSDART00000193254
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr3_-_1146497 | 4.32 |
ENSDART00000149061
|
si:ch73-211l13.2
|
si:ch73-211l13.2 |
| chr23_-_36449111 | 4.21 |
ENSDART00000110478
|
zgc:174906
|
zgc:174906 |
| chr3_-_33437796 | 4.18 |
ENSDART00000075499
|
si:dkey-283b1.7
|
si:dkey-283b1.7 |
| chr1_-_26023678 | 4.07 |
ENSDART00000054202
|
si:ch211-145b13.5
|
si:ch211-145b13.5 |
| chr2_+_36862473 | 3.93 |
ENSDART00000135624
|
si:dkey-193b15.8
|
si:dkey-193b15.8 |
| chr5_-_24231139 | 3.91 |
ENSDART00000143492
|
senp3a
|
SUMO1/sentrin/SMT3 specific peptidase 3a |
| chr7_+_49078890 | 3.77 |
ENSDART00000067857
|
b4galnt4b
|
beta-1,4-N-acetyl-galactosaminyl transferase 4b |
| chr14_-_1958994 | 3.76 |
ENSDART00000161783
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr25_-_12788370 | 3.55 |
ENSDART00000158551
|
slc7a5
|
solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
| chr5_-_38384289 | 3.48 |
ENSDART00000135260
|
mink1
|
misshapen-like kinase 1 |
| chr2_-_24068848 | 3.42 |
ENSDART00000145526
|
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr21_+_36774542 | 3.38 |
ENSDART00000130790
|
prr7
|
proline rich 7 (synaptic) |
| chr10_+_21677058 | 3.33 |
ENSDART00000171499
ENSDART00000157516 |
pcdh1gb2
|
protocadherin 1 gamma b 2 |
| chr17_+_25290136 | 3.33 |
ENSDART00000173295
|
kbtbd11
|
kelch repeat and BTB (POZ) domain containing 11 |
| chr23_-_637347 | 3.23 |
ENSDART00000132175
|
l1camb
|
L1 cell adhesion molecule, paralog b |
| chr16_-_16237844 | 3.18 |
ENSDART00000168747
ENSDART00000111912 |
rbm12b
|
RNA binding motif protein 12B |
| chr8_+_46217861 | 3.16 |
ENSDART00000038790
|
angptl7
|
angiopoietin-like 7 |
| chr7_-_38340674 | 3.16 |
ENSDART00000075782
|
slc7a10a
|
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10a |
| chr2_-_7696503 | 3.14 |
ENSDART00000169709
|
CABZ01055306.1
|
|
| chr2_-_16217344 | 3.13 |
ENSDART00000152031
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr22_-_31517300 | 3.12 |
ENSDART00000164799
|
slc6a6b
|
solute carrier family 6 (neurotransmitter transporter), member 6b |
| chr4_-_68913650 | 3.11 |
ENSDART00000184297
|
si:dkey-264f17.5
|
si:dkey-264f17.5 |
| chr10_-_1718395 | 3.07 |
ENSDART00000137620
|
si:ch73-46j18.5
|
si:ch73-46j18.5 |
| chr7_+_69470442 | 3.03 |
ENSDART00000189593
|
gabarapb
|
GABA(A) receptor-associated protein b |
| chr1_-_44701313 | 2.99 |
ENSDART00000193926
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr10_-_42131408 | 2.79 |
ENSDART00000076693
|
stambpa
|
STAM binding protein a |
| chr13_-_33317323 | 2.77 |
ENSDART00000110295
ENSDART00000144848 ENSDART00000136701 |
tmem234
|
transmembrane protein 234 |
| chr7_+_25003313 | 2.77 |
ENSDART00000131935
|
naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae) |
| chr15_+_26933196 | 2.70 |
ENSDART00000023842
|
ppm1da
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Da |
| chr11_+_37768298 | 2.66 |
ENSDART00000166886
|
sox13
|
SRY (sex determining region Y)-box 13 |
| chr8_-_16697615 | 2.61 |
ENSDART00000187929
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr1_+_33668236 | 2.57 |
ENSDART00000122316
ENSDART00000102184 |
arl13b
|
ADP-ribosylation factor-like 13b |
| chr14_-_8080416 | 2.55 |
ENSDART00000045109
|
zgc:92242
|
zgc:92242 |
| chr4_+_15954293 | 2.54 |
ENSDART00000132695
|
si:dkey-117n7.4
|
si:dkey-117n7.4 |
| chr4_-_13567387 | 2.53 |
ENSDART00000132971
ENSDART00000102010 |
mdm1
|
Mdm1 nuclear protein homolog (mouse) |
| chr1_+_37391141 | 2.43 |
ENSDART00000083593
ENSDART00000168647 |
sparcl1
|
SPARC-like 1 |
| chr8_-_2153147 | 2.41 |
ENSDART00000124093
|
si:dkeyp-117b11.1
|
si:dkeyp-117b11.1 |
| chr18_-_14337450 | 2.35 |
ENSDART00000061435
|
hsbp1b
|
heat shock factor binding protein 1b |
| chr7_-_28565230 | 2.35 |
ENSDART00000028887
|
tmem9b
|
TMEM9 domain family, member B |
| chr17_+_7595356 | 2.33 |
ENSDART00000130625
|
si:dkeyp-110a12.4
|
si:dkeyp-110a12.4 |
| chr9_+_8929764 | 2.32 |
ENSDART00000102562
|
ankrd10b
|
ankyrin repeat domain 10b |
| chr9_+_24920677 | 2.29 |
ENSDART00000037025
|
slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr18_-_46063773 | 2.27 |
ENSDART00000078561
|
si:ch73-262h23.4
|
si:ch73-262h23.4 |
| chr19_+_20708460 | 2.27 |
ENSDART00000034124
|
rab5aa
|
RAB5A, member RAS oncogene family, a |
| chr14_-_2004291 | 2.20 |
ENSDART00000114039
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr17_+_43595692 | 2.17 |
ENSDART00000156271
|
cfap99
|
cilia and flagella associated protein 99 |
| chr14_-_2036604 | 2.16 |
ENSDART00000192446
|
BX005294.2
|
|
| chr17_-_7436766 | 2.12 |
ENSDART00000162002
|
grm1b
|
glutamate receptor, metabotropic 1b |
| chr14_+_6429399 | 2.08 |
ENSDART00000149783
ENSDART00000148461 |
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr23_+_19814371 | 2.08 |
ENSDART00000182897
|
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr14_-_48033073 | 2.08 |
ENSDART00000193115
ENSDART00000169300 ENSDART00000188036 ENSDART00000183432 ENSDART00000180973 |
rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr6_-_13022166 | 2.02 |
ENSDART00000157139
|
tmbim1a
|
transmembrane BAX inhibitor motif containing 1a |
| chr5_-_32363372 | 2.00 |
ENSDART00000098045
|
gas1b
|
growth arrest-specific 1b |
| chr16_+_25343934 | 1.95 |
ENSDART00000109710
|
zfat
|
zinc finger and AT hook domain containing |
| chr1_+_46509176 | 1.95 |
ENSDART00000166028
|
mcf2la
|
mcf.2 cell line derived transforming sequence-like a |
| chr8_+_16990120 | 1.93 |
ENSDART00000018934
|
pde4d
|
phosphodiesterase 4D, cAMP-specific |
| chr1_+_29798140 | 1.88 |
ENSDART00000019743
|
tm9sf2
|
transmembrane 9 superfamily member 2 |
| chr5_-_37875636 | 1.81 |
ENSDART00000184674
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr16_+_32082547 | 1.80 |
ENSDART00000190122
|
prpf31
|
PRP31 pre-mRNA processing factor 31 homolog (yeast) |
| chr16_+_12812472 | 1.79 |
ENSDART00000008535
|
u2af2a
|
U2 small nuclear RNA auxiliary factor 2a |
| chr16_+_32082359 | 1.77 |
ENSDART00000140794
ENSDART00000137029 |
prpf31
|
PRP31 pre-mRNA processing factor 31 homolog (yeast) |
| chr14_-_2318590 | 1.72 |
ENSDART00000192735
|
pcdh2ab8
|
protocadherin 2 alpha b 8 |
| chr6_+_28877306 | 1.71 |
ENSDART00000065137
ENSDART00000123189 ENSDART00000065135 ENSDART00000181512 ENSDART00000130799 |
tp63
|
tumor protein p63 |
| chr6_+_18367388 | 1.70 |
ENSDART00000163394
|
dgke
|
diacylglycerol kinase, epsilon |
| chr10_-_25591194 | 1.58 |
ENSDART00000131640
|
tiam1a
|
T cell lymphoma invasion and metastasis 1a |
| chr7_-_30217000 | 1.52 |
ENSDART00000173646
|
znf710b
|
zinc finger protein 710b |
| chr19_+_22062202 | 1.51 |
ENSDART00000100181
|
sall3b
|
spalt-like transcription factor 3b |
| chr25_+_28825657 | 1.48 |
ENSDART00000153625
|
nfybb
|
nuclear transcription factor Y, beta b |
| chr11_+_7580079 | 1.47 |
ENSDART00000091550
ENSDART00000193223 ENSDART00000193386 |
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr1_-_38361496 | 1.46 |
ENSDART00000015323
|
fbxo8
|
F-box protein 8 |
| chr6_-_41135215 | 1.43 |
ENSDART00000001861
|
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr23_-_7755373 | 1.34 |
ENSDART00000162868
|
PCMTD2 (1 of many)
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
| chr3_-_16110100 | 1.31 |
ENSDART00000051807
|
lasp1
|
LIM and SH3 protein 1 |
| chr23_+_21544227 | 1.30 |
ENSDART00000140253
|
arhgef10lb
|
Rho guanine nucleotide exchange factor (GEF) 10-like b |
| chr6_-_55347616 | 1.28 |
ENSDART00000172081
|
pcif1
|
PDX1 C-terminal inhibiting factor 1 |
| chr13_+_9559461 | 1.28 |
ENSDART00000047740
|
wdr32
|
WD repeat domain 32 |
| chr10_-_32660716 | 1.27 |
ENSDART00000063544
ENSDART00000141013 |
atg101
|
autophagy related 101 |
| chr8_-_16712111 | 1.26 |
ENSDART00000184147
ENSDART00000180419 ENSDART00000076600 |
rpe65c
|
retinal pigment epithelium-specific protein 65c |
| chr3_-_16110351 | 1.25 |
ENSDART00000064838
|
lasp1
|
LIM and SH3 protein 1 |
| chr11_-_21267486 | 1.24 |
ENSDART00000128681
|
DYRK3
|
zgc:172180 |
| chr19_+_20724347 | 1.23 |
ENSDART00000090757
|
kat2b
|
K(lysine) acetyltransferase 2B |
| chr4_-_18939338 | 1.21 |
ENSDART00000132081
ENSDART00000042250 |
rap1b
|
RAP1B, member of RAS oncogene family |
| chr8_-_24252933 | 1.20 |
ENSDART00000057624
|
zgc:110353
|
zgc:110353 |
| chr1_+_26071869 | 1.20 |
ENSDART00000059264
|
mxd4
|
MAX dimerization protein 4 |
| chr19_-_4010263 | 1.17 |
ENSDART00000159605
ENSDART00000165541 |
map7d1b
|
MAP7 domain containing 1b |
| chr6_-_55297274 | 1.16 |
ENSDART00000184283
|
ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr5_-_28149767 | 1.15 |
ENSDART00000051515
|
zgc:110329
|
zgc:110329 |
| chr25_-_35524166 | 1.13 |
ENSDART00000156782
|
slc5a12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr3_-_33175583 | 1.11 |
ENSDART00000126022
|
rarab
|
retinoic acid receptor, alpha b |
| chr20_-_29051696 | 1.10 |
ENSDART00000140350
|
thbs1b
|
thrombospondin 1b |
| chr22_-_16494406 | 1.10 |
ENSDART00000062727
|
stx6
|
syntaxin 6 |
| chr22_-_784110 | 1.06 |
ENSDART00000061775
|
rbbp5
|
retinoblastoma binding protein 5 |
| chr8_+_9149436 | 1.05 |
ENSDART00000064090
|
pnck
|
pregnancy up-regulated nonubiquitous CaM kinase |
| chr25_+_22017182 | 1.02 |
ENSDART00000156517
|
si:dkey-217l24.1
|
si:dkey-217l24.1 |
| chr8_-_52229462 | 1.02 |
ENSDART00000185949
ENSDART00000051825 |
tcf7l1b
|
transcription factor 7 like 1b |
| chr1_+_41487339 | 1.01 |
ENSDART00000142508
ENSDART00000114449 |
si:ch211-89o9.6
|
si:ch211-89o9.6 |
| chr16_+_11242443 | 1.00 |
ENSDART00000024935
|
gsk3ab
|
glycogen synthase kinase 3 alpha b |
| chr15_+_31911989 | 0.94 |
ENSDART00000111472
|
brca2
|
breast cancer 2, early onset |
| chr25_+_25124684 | 0.94 |
ENSDART00000167542
|
ldha
|
lactate dehydrogenase A4 |
| chr24_+_20920563 | 0.92 |
ENSDART00000037224
|
cst14a.2
|
cystatin 14a, tandem duplicate 2 |
| chr6_+_20647155 | 0.91 |
ENSDART00000193477
|
slc19a1
|
solute carrier family 19 (folate transporter), member 1 |
| chr8_-_43456025 | 0.90 |
ENSDART00000001092
ENSDART00000140618 |
ncor2
|
nuclear receptor corepressor 2 |
| chr3_+_61523035 | 0.85 |
ENSDART00000106530
|
znf1004
|
zinc finger protein 1004 |
| chr10_+_43189325 | 0.83 |
ENSDART00000185584
|
vcanb
|
versican b |
| chr9_+_537297 | 0.81 |
ENSDART00000165149
|
CU984600.3
|
|
| chr1_+_45839927 | 0.81 |
ENSDART00000148086
ENSDART00000180413 ENSDART00000048191 ENSDART00000179047 |
map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr8_+_30600386 | 0.78 |
ENSDART00000164976
|
specc1la
|
sperm antigen with calponin homology and coiled-coil domains 1-like a |
| chr1_-_28607353 | 0.76 |
ENSDART00000191210
|
FO907124.1
|
|
| chr10_-_35321625 | 0.74 |
ENSDART00000131268
|
rhbdd2
|
rhomboid domain containing 2 |
| chr1_+_594736 | 0.73 |
ENSDART00000166731
|
jam2a
|
junctional adhesion molecule 2a |
| chr1_+_40613297 | 0.69 |
ENSDART00000040798
ENSDART00000168067 ENSDART00000130490 |
naa15b
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit b |
| chr3_+_40284598 | 0.69 |
ENSDART00000009411
|
bud31
|
BUD31 homolog (S. cerevisiae) |
| chr5_-_30984010 | 0.69 |
ENSDART00000182367
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr22_+_34784075 | 0.67 |
ENSDART00000167538
|
lcor
|
ligand dependent nuclear receptor corepressor |
| chr17_-_21057617 | 0.67 |
ENSDART00000148095
ENSDART00000048853 |
ube2d1a
|
ubiquitin-conjugating enzyme E2D 1a |
| chr25_+_10485103 | 0.61 |
ENSDART00000104576
|
psmd13
|
proteasome 26S subunit, non-ATPase 13 |
| chr20_+_4222357 | 0.61 |
ENSDART00000188331
|
ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr22_-_9861531 | 0.60 |
ENSDART00000193197
|
si:dkey-253d23.2
|
si:dkey-253d23.2 |
| chr22_-_9860792 | 0.58 |
ENSDART00000155908
|
si:dkey-253d23.2
|
si:dkey-253d23.2 |
| chr3_-_60856157 | 0.57 |
ENSDART00000053502
|
CABZ01087513.1
|
|
| chr8_-_16697912 | 0.56 |
ENSDART00000076542
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr15_+_37546391 | 0.56 |
ENSDART00000186625
|
psenen
|
presenilin enhancer gamma secretase subunit |
| chr3_+_41922114 | 0.53 |
ENSDART00000138280
|
lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr17_+_7534365 | 0.52 |
ENSDART00000157123
|
shprh
|
SNF2 histone linker PHD RING helicase |
| chr15_+_35691783 | 0.50 |
ENSDART00000183994
|
CT573366.1
|
|
| chr13_-_12667220 | 0.48 |
ENSDART00000079594
|
fam241a
|
family with sequence similarity 241 member A |
| chr17_-_7218481 | 0.45 |
ENSDART00000181967
|
SAMD5
|
sterile alpha motif domain containing 5 |
| chr23_+_9268083 | 0.42 |
ENSDART00000055054
|
acss2
|
acyl-CoA synthetase short chain family member 2 |
| chr1_-_52790724 | 0.41 |
ENSDART00000139577
ENSDART00000100937 |
patl1
|
protein associated with topoisomerase II homolog 1 (yeast) |
| chr10_+_10972795 | 0.39 |
ENSDART00000127331
|
cdc37l1
|
cell division cycle 37-like 1 |
| chr24_+_36392040 | 0.35 |
ENSDART00000136057
|
arf2b
|
ADP-ribosylation factor 2b |
| chr21_+_22845317 | 0.26 |
ENSDART00000065555
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr17_-_49443247 | 0.26 |
ENSDART00000059245
|
cga
|
glycoprotein hormones, alpha polypeptide |
| chr17_-_49443399 | 0.20 |
ENSDART00000155293
|
cga
|
glycoprotein hormones, alpha polypeptide |
| chr18_+_22138924 | 0.20 |
ENSDART00000183961
|
ripor1
|
RHO family interacting cell polarization regulator 1 |
| chr10_+_3153973 | 0.20 |
ENSDART00000183223
|
hic2
|
hypermethylated in cancer 2 |
| chr17_-_15229787 | 0.17 |
ENSDART00000039165
|
styx
|
serine/threonine/tyrosine interacting protein |
| chr14_+_48960078 | 0.16 |
ENSDART00000165514
|
mif
|
macrophage migration inhibitory factor |
| chr18_+_6479963 | 0.14 |
ENSDART00000092752
ENSDART00000136333 |
wash1
|
WAS protein family homolog 1 |
| chr11_+_24820542 | 0.11 |
ENSDART00000135443
|
kdm5ba
|
lysine (K)-specific demethylase 5Ba |
| chr5_-_30984271 | 0.10 |
ENSDART00000051392
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr7_+_24729558 | 0.08 |
ENSDART00000111542
ENSDART00000170100 |
shroom4
|
shroom family member 4 |
| chr20_+_4221978 | 0.06 |
ENSDART00000171898
|
ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr12_-_22400999 | 0.05 |
ENSDART00000153194
|
si:dkey-38p12.3
|
si:dkey-38p12.3 |
| chr12_+_2428247 | 0.04 |
ENSDART00000152529
|
lrrc18b
|
leucine rich repeat containing 18b |
| chr5_+_45138934 | 0.03 |
ENSDART00000041412
ENSDART00000136002 |
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox12
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 11.2 | GO:0021982 | pineal gland development(GO:0021982) |
| 1.1 | 4.4 | GO:0016116 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 1.1 | 7.6 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 1.1 | 3.2 | GO:0042942 | D-amino acid transport(GO:0042940) D-alanine transport(GO:0042941) D-serine transport(GO:0042942) |
| 0.9 | 9.2 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.9 | 2.6 | GO:1903441 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.8 | 2.3 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.8 | 2.3 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.6 | 3.6 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.5 | 3.8 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.4 | 9.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.3 | 11.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.3 | 0.9 | GO:0015884 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.3 | 4.9 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.2 | 3.5 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.2 | 3.9 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.2 | 7.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.2 | 1.2 | GO:0046929 | negative regulation of neurotransmitter secretion(GO:0046929) response to cAMP(GO:0051591) cellular response to cAMP(GO:0071320) negative regulation of synaptic vesicle transport(GO:1902804) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 | 1.7 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.2 | 2.1 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.2 | 1.0 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.2 | 12.2 | GO:1901214 | regulation of neuron death(GO:1901214) |
| 0.2 | 3.0 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 0.5 | GO:0010893 | positive regulation of steroid biosynthetic process(GO:0010893) positive regulation of steroid metabolic process(GO:0045940) |
| 0.1 | 7.3 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.1 | 3.2 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 2.7 | GO:0046660 | female sex differentiation(GO:0046660) |
| 0.1 | 5.1 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.1 | 3.4 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.1 | 7.0 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 1.3 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 1.9 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 2.8 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 1.1 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.1 | 0.5 | GO:0055016 | hypochord development(GO:0055016) |
| 0.1 | 7.2 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.1 | 2.8 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.1 | 3.1 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.1 | 4.2 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 | 0.8 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 1.7 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 2.3 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 0.4 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.1 | 2.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 1.8 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 0.3 | GO:0060547 | regulation of necrotic cell death(GO:0010939) regulation of necroptotic process(GO:0060544) negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.1 | 15.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.4 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.8 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 1.6 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.8 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 2.0 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.9 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 1.3 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.2 | GO:0043393 | regulation of protein binding(GO:0043393) |
| 0.0 | 1.5 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.7 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.7 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 1.2 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 1.3 | GO:1905037 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 2.3 | GO:1990266 | neutrophil migration(GO:1990266) |
| 0.0 | 0.7 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.4 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 2.6 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 2.7 | GO:0045165 | cell fate commitment(GO:0045165) |
| 0.0 | 6.4 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 21.0 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.8 | 13.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.7 | 5.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.6 | 2.3 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.4 | 3.6 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.3 | 7.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.3 | 3.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 1.8 | GO:0089701 | U2AF(GO:0089701) |
| 0.2 | 3.9 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 0.7 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 1.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 3.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 2.6 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 4.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 3.0 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 2.8 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.1 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 1.1 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 0.5 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 2.1 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 0.6 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 1.3 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 17.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 3.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 3.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 9.2 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 4.0 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.7 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 1.1 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 11.7 | GO:0005794 | Golgi apparatus(GO:0005794) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.4 | GO:0050251 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.8 | 3.8 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.7 | 5.7 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.7 | 2.8 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 0.4 | 11.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.4 | 2.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.3 | 7.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.3 | 1.3 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.3 | 0.9 | GO:0008518 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.3 | 2.1 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.3 | 1.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.3 | 7.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 3.9 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.2 | 9.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 1.8 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.2 | 3.0 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 2.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.2 | 0.8 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.2 | 1.2 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.2 | 6.2 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 3.0 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 2.7 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 0.9 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 4.6 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.1 | 3.6 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.1 | 6.7 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.1 | 3.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.4 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.1 | 1.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 1.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.8 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 1.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 16.3 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 4.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 7.2 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 31.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.2 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 9.7 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.9 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 9.2 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 8.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 1.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.4 | GO:0051087 | chaperone binding(GO:0051087) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 18.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 0.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 3.9 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 2.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 2.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 4.3 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 2.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 3.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 2.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 5.1 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.2 | 1.2 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.2 | 11.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 3.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 1.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 1.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 0.9 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 3.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.2 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.5 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.9 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.8 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |