Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
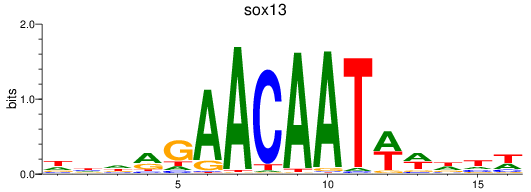
Results for sox13
Z-value: 0.98
Transcription factors associated with sox13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox13
|
ENSDARG00000030297 | SRY-box transcription factor 13 |
|
sox13
|
ENSDARG00000112638 | SRY-box transcription factor 13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox13 | dr11_v1_chr11_+_37803773_37803773 | -0.65 | 1.2e-12 | Click! |
Activity profile of sox13 motif
Sorted Z-values of sox13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_24448278 | 8.17 |
ENSDART00000057584
|
slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr22_-_15602760 | 6.89 |
ENSDART00000009054
|
tpm4a
|
tropomyosin 4a |
| chr21_+_5589923 | 6.85 |
ENSDART00000160885
|
stbd1
|
starch binding domain 1 |
| chr2_+_16780643 | 6.27 |
ENSDART00000125647
ENSDART00000108611 ENSDART00000181245 ENSDART00000163194 |
tfa
|
transferrin-a |
| chr21_+_40092301 | 5.77 |
ENSDART00000145150
|
serpinf2a
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2a |
| chr25_-_19433244 | 5.68 |
ENSDART00000154778
|
map1ab
|
microtubule-associated protein 1Ab |
| chr8_-_32497581 | 4.55 |
ENSDART00000176298
ENSDART00000183340 |
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr6_-_58764672 | 4.39 |
ENSDART00000154322
|
soat2
|
sterol O-acyltransferase 2 |
| chr4_+_17279966 | 3.97 |
ENSDART00000067005
ENSDART00000137487 |
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr22_+_38192568 | 3.81 |
ENSDART00000126323
|
cp
|
ceruloplasmin |
| chr8_-_32497815 | 3.74 |
ENSDART00000122359
|
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr25_-_4146947 | 3.73 |
ENSDART00000129268
|
fads2
|
fatty acid desaturase 2 |
| chr15_-_21155641 | 3.49 |
ENSDART00000061098
ENSDART00000046443 |
A2ML1 (1 of many)
|
si:dkey-105h12.2 |
| chr22_-_17606575 | 3.49 |
ENSDART00000183951
|
gpx4a
|
glutathione peroxidase 4a |
| chr9_-_16109001 | 3.33 |
ENSDART00000053473
|
upp2
|
uridine phosphorylase 2 |
| chr1_+_12231478 | 3.33 |
ENSDART00000111485
|
tmod1
|
tropomodulin 1 |
| chr20_+_10538025 | 3.33 |
ENSDART00000129762
|
serpina1l
|
serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1, like |
| chr11_-_23458792 | 3.28 |
ENSDART00000032844
|
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr23_+_22200467 | 3.12 |
ENSDART00000025414
|
slc2a1a
|
solute carrier family 2 (facilitated glucose transporter), member 1a |
| chr2_+_26240631 | 3.11 |
ENSDART00000129895
|
palm1b
|
paralemmin 1b |
| chr21_-_13123176 | 3.07 |
ENSDART00000144866
ENSDART00000024616 |
fam219aa
|
family with sequence similarity 219, member Aa |
| chr17_-_33289304 | 3.06 |
ENSDART00000135118
ENSDART00000040346 |
efr3ba
|
EFR3 homolog Ba (S. cerevisiae) |
| chr23_-_26521970 | 3.00 |
ENSDART00000143712
|
si:dkey-205h13.1
|
si:dkey-205h13.1 |
| chr12_-_6177894 | 2.95 |
ENSDART00000152292
|
a1cf
|
apobec1 complementation factor |
| chr20_-_27225876 | 2.73 |
ENSDART00000149204
ENSDART00000149732 |
si:dkey-85n7.7
|
si:dkey-85n7.7 |
| chr23_-_26522760 | 2.69 |
ENSDART00000142417
ENSDART00000135606 ENSDART00000122668 |
si:dkey-205h13.1
|
si:dkey-205h13.1 |
| chr24_+_22485710 | 2.66 |
ENSDART00000146058
|
si:dkey-40h20.1
|
si:dkey-40h20.1 |
| chr13_-_31397987 | 2.55 |
ENSDART00000008287
|
pgam1a
|
phosphoglycerate mutase 1a |
| chr14_-_33454595 | 2.51 |
ENSDART00000109615
ENSDART00000173267 ENSDART00000185737 ENSDART00000190989 |
tmem255a
|
transmembrane protein 255A |
| chr16_-_43971258 | 2.44 |
ENSDART00000141941
|
zfpm2a
|
zinc finger protein, FOG family member 2a |
| chr14_+_34486629 | 2.40 |
ENSDART00000131861
|
tmsb2
|
thymosin beta 2 |
| chr16_+_44298902 | 2.40 |
ENSDART00000114795
|
dpys
|
dihydropyrimidinase |
| chr14_-_24251057 | 2.37 |
ENSDART00000114169
|
bnip1a
|
BCL2 interacting protein 1a |
| chr14_+_30285613 | 2.34 |
ENSDART00000173090
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr20_-_40451115 | 2.33 |
ENSDART00000075092
|
pkib
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr19_-_20114149 | 2.31 |
ENSDART00000052620
|
npy
|
neuropeptide Y |
| chr18_+_17428506 | 2.28 |
ENSDART00000100223
|
zgc:91860
|
zgc:91860 |
| chr22_+_1887750 | 2.21 |
ENSDART00000180917
|
si:dkey-15h8.17
|
si:dkey-15h8.17 |
| chr18_+_17428258 | 2.20 |
ENSDART00000010452
|
zgc:91860
|
zgc:91860 |
| chr9_+_50316921 | 2.19 |
ENSDART00000098687
|
GRB14
|
growth factor receptor bound protein 14 |
| chr3_-_8246309 | 2.14 |
ENSDART00000134497
ENSDART00000015232 |
cyp2k22
|
cytochrome P450, family 2, subfamily K, polypeptide 22 |
| chr17_+_30522764 | 2.09 |
ENSDART00000168051
ENSDART00000193728 |
nhsl1a
|
NHS-like 1a |
| chr21_+_38817785 | 2.07 |
ENSDART00000177616
ENSDART00000149085 |
hnf1bb
|
HNF1 homeobox Bb |
| chr19_-_20113696 | 2.05 |
ENSDART00000188813
|
npy
|
neuropeptide Y |
| chr17_-_25737452 | 2.04 |
ENSDART00000152021
|
si:ch211-214p16.3
|
si:ch211-214p16.3 |
| chr9_+_22359919 | 2.03 |
ENSDART00000009591
|
crygs4
|
crystallin, gamma S4 |
| chr19_-_3931917 | 1.99 |
ENSDART00000162532
|
map7d1b
|
MAP7 domain containing 1b |
| chr8_+_30664077 | 1.97 |
ENSDART00000138750
|
adora2aa
|
adenosine A2a receptor a |
| chr9_+_2574122 | 1.96 |
ENSDART00000166326
ENSDART00000191822 |
CIR1
|
si:ch73-167c12.2 |
| chr7_-_35516251 | 1.95 |
ENSDART00000045628
|
irx6a
|
iroquois homeobox 6a |
| chr21_-_2261720 | 1.91 |
ENSDART00000170161
|
si:ch73-299h12.2
|
si:ch73-299h12.2 |
| chr16_-_17197546 | 1.90 |
ENSDART00000139939
ENSDART00000135146 ENSDART00000063800 ENSDART00000163606 |
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr2_+_37875789 | 1.88 |
ENSDART00000036318
ENSDART00000127679 |
cbln13
|
cerebellin 13 |
| chr13_-_8692860 | 1.87 |
ENSDART00000058107
|
mcfd2
|
multiple coagulation factor deficiency 2 |
| chr11_-_1509773 | 1.87 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr2_+_50626476 | 1.86 |
ENSDART00000018150
|
neurod6b
|
neuronal differentiation 6b |
| chr20_+_43942278 | 1.86 |
ENSDART00000100571
|
clic5b
|
chloride intracellular channel 5b |
| chr22_-_15602593 | 1.85 |
ENSDART00000036075
|
tpm4a
|
tropomyosin 4a |
| chr20_-_29475172 | 1.84 |
ENSDART00000183164
|
scg5
|
secretogranin V |
| chr19_-_34873566 | 1.82 |
ENSDART00000016057
|
ctnnal1
|
catenin (cadherin-associated protein), alpha-like 1 |
| chr18_+_13837746 | 1.82 |
ENSDART00000169552
|
cdh13
|
cadherin 13, H-cadherin (heart) |
| chr24_+_17269849 | 1.81 |
ENSDART00000017605
|
spag6
|
sperm associated antigen 6 |
| chr10_+_2587234 | 1.79 |
ENSDART00000126937
|
wu:fb59d01
|
wu:fb59d01 |
| chr7_+_21859337 | 1.79 |
ENSDART00000159626
|
si:dkey-85k7.7
|
si:dkey-85k7.7 |
| chr24_+_15897717 | 1.79 |
ENSDART00000105956
|
neto1l
|
neuropilin (NRP) and tolloid (TLL)-like 1, like |
| chr19_-_45960191 | 1.78 |
ENSDART00000052434
ENSDART00000172732 |
eif3hb
|
eukaryotic translation initiation factor 3, subunit H, b |
| chr2_+_23007675 | 1.78 |
ENSDART00000163649
|
mknk2a
|
MAP kinase interacting serine/threonine kinase 2a |
| chr12_-_35988586 | 1.77 |
ENSDART00000157746
|
pde6gb
|
phosphodiesterase 6G, cGMP-specific, rod, gamma, paralog b |
| chr16_-_563732 | 1.77 |
ENSDART00000183394
|
irx2a
|
iroquois homeobox 2a |
| chr5_-_3627110 | 1.76 |
ENSDART00000156071
|
si:zfos-375h5.1
|
si:zfos-375h5.1 |
| chr8_+_40476811 | 1.74 |
ENSDART00000129772
|
gck
|
glucokinase (hexokinase 4) |
| chr14_-_24111292 | 1.72 |
ENSDART00000186611
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr22_-_18112374 | 1.71 |
ENSDART00000191154
|
ncanb
|
neurocan b |
| chr18_-_21188768 | 1.69 |
ENSDART00000060166
|
got2a
|
glutamic-oxaloacetic transaminase 2a, mitochondrial |
| chr16_-_16212615 | 1.69 |
ENSDART00000059905
|
upp1
|
uridine phosphorylase 1 |
| chr12_+_28574863 | 1.67 |
ENSDART00000153284
|
tbkbp1
|
TBK1 binding protein 1 |
| chr20_-_15089738 | 1.66 |
ENSDART00000164552
|
si:dkey-239i20.2
|
si:dkey-239i20.2 |
| chr20_-_29474859 | 1.65 |
ENSDART00000152906
ENSDART00000045249 |
scg5
|
secretogranin V |
| chr25_-_35360096 | 1.65 |
ENSDART00000154053
ENSDART00000171917 |
si:ch73-147o17.1
|
si:ch73-147o17.1 |
| chr12_-_20584413 | 1.62 |
ENSDART00000170923
|
FP885542.2
|
|
| chr7_-_32980017 | 1.61 |
ENSDART00000113744
|
pkp3b
|
plakophilin 3b |
| chr16_+_6336952 | 1.61 |
ENSDART00000148855
|
cmtm8b
|
CKLF-like MARVEL transmembrane domain containing 8b |
| chr7_-_51757085 | 1.59 |
ENSDART00000167747
|
leap2
|
liver-expressed antimicrobial peptide 2 |
| chr19_-_15397946 | 1.57 |
ENSDART00000143480
|
hivep3a
|
human immunodeficiency virus type I enhancer binding protein 3a |
| chr10_+_39263827 | 1.56 |
ENSDART00000172509
|
foxred1
|
FAD-dependent oxidoreductase domain containing 1 |
| chr14_+_22132388 | 1.55 |
ENSDART00000109065
|
ccng1
|
cyclin G1 |
| chr13_+_18354670 | 1.55 |
ENSDART00000023806
ENSDART00000133931 |
zgc:110319
|
zgc:110319 |
| chr2_+_3502430 | 1.55 |
ENSDART00000113395
|
SERP1
|
zgc:92744 |
| chr4_+_75200467 | 1.55 |
ENSDART00000122593
|
CABZ01043955.1
|
|
| chr14_-_25599002 | 1.54 |
ENSDART00000040955
|
slc25a48
|
solute carrier family 25, member 48 |
| chr12_+_31729498 | 1.52 |
ENSDART00000188546
ENSDART00000182562 ENSDART00000186147 |
RNF157
|
si:dkey-49c17.3 |
| chr3_+_12835370 | 1.51 |
ENSDART00000166089
|
si:ch211-8c17.3
|
si:ch211-8c17.3 |
| chr24_-_7321928 | 1.51 |
ENSDART00000167570
ENSDART00000045150 |
actr3b
|
ARP3 actin related protein 3 homolog B |
| chr1_+_26445615 | 1.50 |
ENSDART00000180810
|
g3bp2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr7_-_52096498 | 1.50 |
ENSDART00000098688
ENSDART00000098690 |
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr13_+_31497236 | 1.48 |
ENSDART00000146752
|
lrrc9
|
leucine rich repeat containing 9 |
| chr6_+_39493864 | 1.48 |
ENSDART00000086263
|
mettl7a
|
methyltransferase like 7A |
| chr23_+_36771593 | 1.48 |
ENSDART00000078240
|
march9
|
membrane-associated ring finger (C3HC4) 9 |
| chr6_+_39905021 | 1.48 |
ENSDART00000064904
|
endou
|
endonuclease, polyU-specific |
| chr16_-_44127307 | 1.48 |
ENSDART00000104583
ENSDART00000151936 ENSDART00000058685 ENSDART00000190830 |
zfpm2a
|
zinc finger protein, FOG family member 2a |
| chr7_+_58686860 | 1.46 |
ENSDART00000052332
|
penkb
|
proenkephalin b |
| chr11_-_29623380 | 1.45 |
ENSDART00000162587
ENSDART00000193935 ENSDART00000191646 |
chd5
|
chromodomain helicase DNA binding protein 5 |
| chr4_+_72578548 | 1.44 |
ENSDART00000174035
|
si:cabz01054394.5
|
si:cabz01054394.5 |
| chr8_-_8698607 | 1.43 |
ENSDART00000046712
|
zgc:86609
|
zgc:86609 |
| chr5_+_9417409 | 1.42 |
ENSDART00000125421
ENSDART00000130265 |
ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr10_+_21677058 | 1.42 |
ENSDART00000171499
ENSDART00000157516 |
pcdh1gb2
|
protocadherin 1 gamma b 2 |
| chr11_+_23760470 | 1.42 |
ENSDART00000175688
ENSDART00000121874 ENSDART00000086720 |
nfasca
|
neurofascin homolog (chicken) a |
| chr14_-_41308561 | 1.41 |
ENSDART00000138232
|
arl13a
|
ADP-ribosylation factor-like 13A |
| chr24_+_20325312 | 1.41 |
ENSDART00000007997
|
acaa1
|
acetyl-CoA acyltransferase 1 |
| chr13_+_25720969 | 1.40 |
ENSDART00000046050
|
pcbd1
|
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha |
| chr16_-_43011470 | 1.39 |
ENSDART00000131898
ENSDART00000142003 ENSDART00000017966 |
nudt17
|
nudix (nucleoside diphosphate linked moiety X)-type motif 17 |
| chr20_+_34029820 | 1.38 |
ENSDART00000143901
ENSDART00000134305 |
prg4b
|
proteoglycan 4b |
| chr6_-_43922813 | 1.37 |
ENSDART00000123341
|
prok2
|
prokineticin 2 |
| chr11_-_25733910 | 1.36 |
ENSDART00000171935
|
brpf3a
|
bromodomain and PHD finger containing, 3a |
| chr18_-_19405616 | 1.35 |
ENSDART00000191290
ENSDART00000090855 |
megf11
|
multiple EGF-like-domains 11 |
| chr12_+_33395748 | 1.34 |
ENSDART00000129458
|
fasn
|
fatty acid synthase |
| chr2_+_34967022 | 1.33 |
ENSDART00000134926
|
astn1
|
astrotactin 1 |
| chr14_+_26247319 | 1.33 |
ENSDART00000192793
|
CCDC69
|
coiled-coil domain containing 69 |
| chr3_-_18710009 | 1.33 |
ENSDART00000142478
|
grid2ipa
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, a |
| chr24_-_17067284 | 1.32 |
ENSDART00000111237
|
armc3
|
armadillo repeat containing 3 |
| chr7_+_60079302 | 1.32 |
ENSDART00000051524
|
etnppl
|
ethanolamine-phosphate phospho-lyase |
| chr5_+_72087619 | 1.32 |
ENSDART00000062885
|
oxt
|
oxytocin |
| chr7_+_37359004 | 1.32 |
ENSDART00000192134
ENSDART00000189239 |
sall1a
|
spalt-like transcription factor 1a |
| chr14_-_45967981 | 1.30 |
ENSDART00000188062
|
macrod1
|
MACRO domain containing 1 |
| chr14_-_2202652 | 1.30 |
ENSDART00000171316
|
si:dkey-262j3.7
|
si:dkey-262j3.7 |
| chr7_+_58699718 | 1.30 |
ENSDART00000049264
|
sdr16c5b
|
short chain dehydrogenase/reductase family 16C, member 5b |
| chr4_-_71006506 | 1.30 |
ENSDART00000172163
|
si:dkeyp-80d11.4
|
si:dkeyp-80d11.4 |
| chr24_+_33589667 | 1.29 |
ENSDART00000152097
|
dnah5l
|
dynein, axonemal, heavy chain 5 like |
| chr22_+_3238474 | 1.29 |
ENSDART00000157954
|
si:ch1073-178p5.3
|
si:ch1073-178p5.3 |
| chr7_-_29292206 | 1.29 |
ENSDART00000086753
|
dapk2a
|
death-associated protein kinase 2a |
| chr23_-_18707418 | 1.28 |
ENSDART00000144668
ENSDART00000141205 ENSDART00000016765 |
zgc:103759
|
zgc:103759 |
| chr13_-_31470439 | 1.28 |
ENSDART00000076574
|
rtn1a
|
reticulon 1a |
| chr16_+_40024883 | 1.28 |
ENSDART00000110100
|
hint3
|
histidine triad nucleotide binding protein 3 |
| chr22_-_6562618 | 1.27 |
ENSDART00000106100
|
zgc:171490
|
zgc:171490 |
| chr1_+_54908895 | 1.26 |
ENSDART00000145652
|
golga7ba
|
golgin A7 family, member Ba |
| chr3_+_25503442 | 1.26 |
ENSDART00000190117
|
mchr1b
|
melanin-concentrating hormone receptor 1b |
| chr18_-_17399291 | 1.25 |
ENSDART00000192075
ENSDART00000060949 ENSDART00000188506 |
zfpm1
|
zinc finger protein, FOG family member 1 |
| chr25_+_19522954 | 1.24 |
ENSDART00000006512
ENSDART00000184279 |
sh3gl3b
|
SH3-domain GRB2-like 3b |
| chr22_-_20950448 | 1.22 |
ENSDART00000002029
|
fkbp8
|
FK506 binding protein 8 |
| chr10_-_7857494 | 1.22 |
ENSDART00000143215
|
inpp5ja
|
inositol polyphosphate-5-phosphatase Ja |
| chr16_+_13855039 | 1.21 |
ENSDART00000113764
ENSDART00000143983 |
zgc:174888
|
zgc:174888 |
| chr24_+_26017094 | 1.21 |
ENSDART00000137851
|
tfr1b
|
transferrin receptor 1b |
| chr14_+_44545092 | 1.21 |
ENSDART00000175454
|
lingo2a
|
leucine rich repeat and Ig domain containing 2a |
| chr1_-_19079957 | 1.20 |
ENSDART00000141795
|
phox2ba
|
paired-like homeobox 2ba |
| chr6_-_15639087 | 1.20 |
ENSDART00000128940
ENSDART00000183092 |
mlpha
|
melanophilin a |
| chr17_-_23412705 | 1.19 |
ENSDART00000126995
|
si:ch211-149k12.3
|
si:ch211-149k12.3 |
| chr8_-_41279326 | 1.18 |
ENSDART00000075491
|
pop5
|
POP5 homolog, ribonuclease P/MRP subunit |
| chr6_-_48473694 | 1.16 |
ENSDART00000154237
|
ppm1j
|
protein phosphatase, Mg2+/Mn2+ dependent, 1J |
| chr24_+_17270129 | 1.16 |
ENSDART00000186729
|
spag6
|
sperm associated antigen 6 |
| chr20_+_20726231 | 1.16 |
ENSDART00000147112
|
zgc:193541
|
zgc:193541 |
| chr19_-_20388064 | 1.16 |
ENSDART00000132272
|
rftn1a
|
raftlin, lipid raft linker 1a |
| chr7_+_5920665 | 1.15 |
ENSDART00000173208
|
si:dkey-23a13.11
|
si:dkey-23a13.11 |
| chr24_+_39277043 | 1.15 |
ENSDART00000165458
|
MPRIP
|
si:ch73-103b11.2 |
| chr21_-_42007213 | 1.15 |
ENSDART00000188804
ENSDART00000092821 ENSDART00000165743 |
gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr12_+_33396489 | 1.14 |
ENSDART00000149960
|
fasn
|
fatty acid synthase |
| chr10_+_21559605 | 1.14 |
ENSDART00000123648
ENSDART00000108584 |
pcdh1a3
pcdh1a3
|
protocadherin 1 alpha 3 protocadherin 1 alpha 3 |
| chr7_+_25033924 | 1.13 |
ENSDART00000170873
|
sb:cb1058
|
sb:cb1058 |
| chr8_+_1839695 | 1.13 |
ENSDART00000148254
ENSDART00000143473 |
snap29
|
synaptosomal-associated protein 29 |
| chr1_-_10647484 | 1.13 |
ENSDART00000164541
ENSDART00000188958 ENSDART00000190904 |
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr21_-_8513192 | 1.13 |
ENSDART00000084378
|
crb2a
|
crumbs family member 2a |
| chr6_+_612594 | 1.13 |
ENSDART00000150903
|
kynu
|
kynureninase |
| chr2_-_9646857 | 1.11 |
ENSDART00000056901
|
zgc:153615
|
zgc:153615 |
| chr4_+_38001640 | 1.11 |
ENSDART00000131318
|
znf1051
|
zinc finger protein 1051 |
| chr4_+_39742119 | 1.11 |
ENSDART00000176004
|
CR749167.2
|
|
| chr3_-_37759969 | 1.10 |
ENSDART00000151105
ENSDART00000151208 |
si:dkey-260c8.6
|
si:dkey-260c8.6 |
| chr2_-_5071888 | 1.10 |
ENSDART00000185036
|
dlg1l
|
discs, large (Drosophila) homolog 1, like |
| chr18_+_1837668 | 1.10 |
ENSDART00000164210
|
CABZ01079192.1
|
|
| chr15_+_23911655 | 1.10 |
ENSDART00000156304
|
si:ch1073-145m9.1
|
si:ch1073-145m9.1 |
| chr5_+_44944778 | 1.09 |
ENSDART00000130428
ENSDART00000044361 ENSDART00000128825 ENSDART00000124637 ENSDART00000126066 ENSDART00000177635 |
dmrt1
|
doublesex and mab-3 related transcription factor 1 |
| chr18_+_21273749 | 1.09 |
ENSDART00000143265
|
hydin
|
HYDIN, axonemal central pair apparatus protein |
| chr23_-_25126003 | 1.09 |
ENSDART00000034953
|
idh3g
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr20_+_38724575 | 1.08 |
ENSDART00000015095
ENSDART00000152972 |
uts1
|
urotensin 1 |
| chr11_-_42752884 | 1.06 |
ENSDART00000186025
|
si:ch73-106k19.5
|
si:ch73-106k19.5 |
| chr22_-_12746539 | 1.05 |
ENSDART00000175374
|
plcd4a
|
phospholipase C, delta 4a |
| chr19_+_23919096 | 1.05 |
ENSDART00000090200
|
snapin
|
SNAP-associated protein |
| chr2_+_31833997 | 1.05 |
ENSDART00000066788
|
epdr1
|
ependymin related 1 |
| chr23_+_31107685 | 1.04 |
ENSDART00000103448
|
tbx18
|
T-box 18 |
| chr19_-_31341850 | 1.04 |
ENSDART00000040810
|
arl4ab
|
ADP-ribosylation factor-like 4ab |
| chr3_-_37759373 | 1.04 |
ENSDART00000150962
|
si:dkey-260c8.6
|
si:dkey-260c8.6 |
| chr6_-_48473395 | 1.04 |
ENSDART00000185096
|
ppm1j
|
protein phosphatase, Mg2+/Mn2+ dependent, 1J |
| chr13_+_8693410 | 1.04 |
ENSDART00000138448
|
ttc7a
|
tetratricopeptide repeat domain 7A |
| chr21_+_7188943 | 1.03 |
ENSDART00000172174
|
agpat2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 (lysophosphatidic acid acyltransferase, beta) |
| chr23_-_45111637 | 1.03 |
ENSDART00000179776
|
si:ch73-168d20.1
|
si:ch73-168d20.1 |
| chr13_-_27675212 | 1.03 |
ENSDART00000141035
|
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr4_-_58846245 | 1.03 |
ENSDART00000170777
|
si:dkey-28i19.1
|
si:dkey-28i19.1 |
| chr10_-_26738209 | 1.03 |
ENSDART00000188590
|
fgf13b
|
fibroblast growth factor 13b |
| chr7_-_51953807 | 1.02 |
ENSDART00000174102
ENSDART00000145645 ENSDART00000052054 |
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr4_+_21129752 | 1.02 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr5_+_58492699 | 1.01 |
ENSDART00000181584
|
FQ378016.1
|
|
| chr4_+_29206813 | 1.01 |
ENSDART00000131893
|
si:dkey-23a23.1
|
si:dkey-23a23.1 |
| chr8_+_32389838 | 1.00 |
ENSDART00000076350
ENSDART00000146901 |
mthfd2l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2 like |
| chr5_+_29652198 | 0.99 |
ENSDART00000184083
|
tsc1a
|
TSC complex subunit 1a |
| chr23_-_24547564 | 0.99 |
ENSDART00000134410
|
si:dkey-31b16.7
|
si:dkey-31b16.7 |
| chr21_-_42007482 | 0.98 |
ENSDART00000075740
|
gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr19_-_7406933 | 0.98 |
ENSDART00000151137
|
oxr1b
|
oxidation resistance 1b |
| chr2_-_44183613 | 0.97 |
ENSDART00000079596
|
cadm3
|
cell adhesion molecule 3 |
| chr15_-_34326461 | 0.97 |
ENSDART00000156682
|
dgkb
|
diacylglycerol kinase, beta |
| chr7_-_6279138 | 0.96 |
ENSDART00000173299
|
si:ch211-220f21.2
|
si:ch211-220f21.2 |
| chr9_-_12443726 | 0.96 |
ENSDART00000102434
|
ehhadh
|
enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase |
| chr19_-_44801918 | 0.95 |
ENSDART00000108507
|
CSMD3 (1 of many)
|
si:ch211-233f16.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox13
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.4 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 1.0 | 4.0 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.6 | 6.4 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.5 | 1.6 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.5 | 2.1 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.5 | 5.0 | GO:0032262 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) |
| 0.5 | 2.0 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.5 | 4.4 | GO:0034433 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.5 | 1.4 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.5 | 1.4 | GO:0072526 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.4 | 2.1 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.4 | 2.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.4 | 3.1 | GO:0046323 | glucose import(GO:0046323) |
| 0.4 | 2.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.4 | 1.5 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.4 | 1.1 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.4 | 2.6 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.4 | 1.1 | GO:0030238 | male sex determination(GO:0030238) |
| 0.4 | 1.1 | GO:1904088 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.3 | 0.9 | GO:1902767 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.3 | 1.2 | GO:0033572 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.3 | 1.2 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.3 | 1.8 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.3 | 0.8 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.3 | 7.5 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.3 | 1.3 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.2 | 1.0 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.2 | 1.8 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.2 | 1.1 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.2 | 0.7 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.2 | 5.8 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.2 | 1.6 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 2.1 | GO:0072098 | anterior/posterior pattern specification involved in pronephros development(GO:0034672) anterior/posterior pattern specification involved in kidney development(GO:0072098) |
| 0.2 | 0.6 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.2 | 0.8 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.2 | 4.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.2 | 1.7 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.2 | 1.1 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.2 | 1.8 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.2 | 2.4 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.2 | 0.5 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.2 | 1.4 | GO:0030431 | sleep(GO:0030431) |
| 0.2 | 0.7 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 | 0.5 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.2 | 0.3 | GO:0042755 | eating behavior(GO:0042755) |
| 0.2 | 0.9 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.2 | 0.6 | GO:1903817 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.2 | 0.8 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.2 | 0.5 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.2 | 1.2 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.2 | 1.2 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.1 | 0.6 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.1 | 0.4 | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.1 | 0.7 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 0.3 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.1 | 1.4 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 0.7 | GO:0070973 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 1.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 2.4 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.1 | 1.7 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.1 | 0.9 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 0.6 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.1 | 1.0 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.1 | 0.7 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 0.7 | GO:0033986 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.1 | 0.5 | GO:0046551 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.1 | 0.6 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.1 | 0.9 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.1 | 2.0 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 1.3 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 0.3 | GO:1901376 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.1 | 1.5 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 1.4 | GO:0042537 | benzene-containing compound metabolic process(GO:0042537) |
| 0.1 | 2.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.3 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 9.1 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 1.8 | GO:0035778 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) |
| 0.1 | 0.7 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 1.4 | GO:0035545 | determination of left/right asymmetry in diencephalon(GO:0035462) determination of left/right asymmetry in nervous system(GO:0035545) |
| 0.1 | 0.4 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) regulation of odontogenesis(GO:0042481) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) |
| 0.1 | 0.3 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.1 | 1.1 | GO:0008105 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.1 | 0.2 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 1.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.4 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.1 | 0.9 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 0.2 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.1 | 0.6 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 2.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 2.1 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 0.8 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.7 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 1.5 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 0.6 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 0.7 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.1 | 2.9 | GO:0046883 | regulation of hormone secretion(GO:0046883) |
| 0.1 | 4.8 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.1 | 1.6 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 1.3 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 1.0 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.5 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.2 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 0.7 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.9 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 1.0 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 0.6 | GO:0060004 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.1 | 0.4 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.1 | 1.0 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.1 | 0.7 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 0.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 0.8 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.2 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 0.5 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.4 | GO:0030262 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.1 | 0.4 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.1 | 0.4 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 | 0.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.8 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 1.1 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.4 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.2 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.8 | GO:0060324 | face development(GO:0060324) |
| 0.0 | 0.4 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.6 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.5 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.6 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.6 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 1.0 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 4.8 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.1 | GO:0016093 | polyprenol metabolic process(GO:0016093) |
| 0.0 | 1.8 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.0 | 0.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 0.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.6 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.0 | 4.3 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.7 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.3 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.4 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.8 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 3.5 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 1.5 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.0 | 0.5 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 1.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.5 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.6 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.4 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.6 | GO:1990089 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.0 | 1.6 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.8 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.2 | GO:0000022 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.3 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 3.6 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.6 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 2.5 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.5 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.2 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 1.0 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.7 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 1.4 | GO:0002455 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 1.0 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 1.4 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.2 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.4 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.0 | 0.4 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.2 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) cortisol metabolic process(GO:0034650) |
| 0.0 | 0.8 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 1.2 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.2 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 1.2 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.4 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.4 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.6 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.6 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 2.3 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.2 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.0 | 0.8 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.9 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 0.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 3.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0032264 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.2 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.2 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.0 | 0.4 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.3 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.7 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.2 | GO:1902307 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.1 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 4.0 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.3 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.3 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.5 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.3 | 1.8 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 0.7 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.2 | 0.9 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.2 | 0.9 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.2 | 1.0 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.2 | 1.0 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.2 | 1.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 2.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 0.9 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.8 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 1.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.6 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 1.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.6 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 1.4 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 0.4 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 1.5 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 1.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 0.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 1.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 1.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 1.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 6.3 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 6.7 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 0.7 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 4.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 8.2 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 6.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 1.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 1.0 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.6 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.3 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 3.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.4 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.0 | 0.2 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.5 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 4.7 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.3 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 2.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.7 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.0 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 0.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 1.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 1.0 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.3 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.2 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.2 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 1.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 26.0 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.4 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 1.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.0 | 2.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.7 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.1 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.3 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.9 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.2 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 6.9 | GO:2001070 | starch binding(GO:2001070) |
| 1.1 | 4.4 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 1.0 | 4.0 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.9 | 4.4 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.7 | 3.5 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.5 | 2.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.5 | 2.5 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.5 | 1.9 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.4 | 1.3 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.4 | 1.7 | GO:0030586 | [methionine synthase] reductase activity(GO:0030586) |
| 0.4 | 1.3 | GO:0030273 | melanin-concentrating hormone receptor activity(GO:0030273) |
| 0.4 | 2.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.4 | 1.5 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.4 | 1.4 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.3 | 5.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 1.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.3 | 0.9 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.3 | 1.2 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.3 | 1.2 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.3 | 1.7 | GO:0005536 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.3 | 0.9 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.3 | 2.0 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.3 | 1.4 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.2 | 6.2 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.2 | 1.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.2 | 0.7 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.2 | 1.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.2 | 0.6 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.2 | 2.4 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.2 | 1.7 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 0.7 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.2 | 2.6 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.2 | 2.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.2 | 1.2 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.1 | 1.3 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 2.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 1.0 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.1 | 1.8 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 0.8 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.7 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.8 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 2.1 | GO:0015145 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.1 | 1.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 0.5 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.5 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 1.5 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 1.0 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.1 | 1.5 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 1.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.5 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.6 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 1.8 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.1 | 0.6 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 0.3 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 0.6 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 1.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.7 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 0.5 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 0.3 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.1 | 9.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.7 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 0.7 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 1.0 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.7 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 0.9 | GO:0070628 | lysine-acetylated histone binding(GO:0070577) proteasome binding(GO:0070628) |
| 0.1 | 0.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 4.7 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 2.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.4 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.1 | 1.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 4.8 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.1 | 1.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.6 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.1 | 0.6 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.4 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.4 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.8 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 3.3 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 1.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.6 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 3.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.4 | GO:0008506 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 8.2 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 2.8 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.6 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.5 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.5 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.6 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.5 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.6 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.7 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.2 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 1.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.3 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.3 | GO:0005549 | olfactory receptor activity(GO:0004984) odorant binding(GO:0005549) |
| 0.0 | 1.1 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.0 | 0.8 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 1.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.4 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 1.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 1.1 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.6 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 3.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 2.0 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.3 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 1.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 1.2 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.4 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 1.3 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.7 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.1 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.0 | 8.2 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.7 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.3 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.3 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.5 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 3.9 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.6 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 1.5 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 1.8 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 0.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 2.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 1.3 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.0 | 0.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.4 | GO:0019902 | phosphatase binding(GO:0019902) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 5.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 5.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 0.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 0.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 2.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 0.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 2.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 4.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.2 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.6 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 7.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.5 | 5.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.3 | 8.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 4.3 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.2 | 4.0 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 2.5 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.1 | 2.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 0.9 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 0.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 2.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 6.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 2.0 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 2.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.6 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 1.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 1.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.7 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 1.1 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 0.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.0 | 0.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |