Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
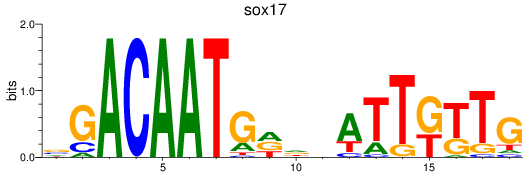
Results for sox17
Z-value: 0.68
Transcription factors associated with sox17
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox17
|
ENSDARG00000101717 | SRY-box transcription factor 17 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox17 | dr11_v1_chr7_-_58776400_58776400 | -0.29 | 5.0e-03 | Click! |
Activity profile of sox17 motif
Sorted Z-values of sox17 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_36738069 | 6.98 |
ENSDART00000105590
|
tdo2a
|
tryptophan 2,3-dioxygenase a |
| chr5_+_37978501 | 6.12 |
ENSDART00000012050
|
apoa1a
|
apolipoprotein A-Ia |
| chr22_-_26175237 | 5.98 |
ENSDART00000108737
|
c3b.2
|
complement component c3b, tandem duplicate 2 |
| chr22_-_26236188 | 5.91 |
ENSDART00000162640
ENSDART00000167169 ENSDART00000138595 |
c3b.1
|
complement component c3b, tandem duplicate 1 |
| chr7_+_21841037 | 4.96 |
ENSDART00000077503
|
tm4sf5
|
transmembrane 4 L six family member 5 |
| chr19_+_9305964 | 4.42 |
ENSDART00000136241
|
si:ch73-15n24.1
|
si:ch73-15n24.1 |
| chr6_-_40058686 | 3.65 |
ENSDART00000103240
|
uroc1
|
urocanate hydratase 1 |
| chr16_-_28658341 | 3.41 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr9_+_34127005 | 3.40 |
ENSDART00000167384
ENSDART00000078065 |
f5
|
coagulation factor V |
| chr18_+_1837668 | 3.12 |
ENSDART00000164210
|
CABZ01079192.1
|
|
| chr21_+_26748141 | 2.99 |
ENSDART00000169025
|
pcxa
|
pyruvate carboxylase a |
| chr2_+_3472832 | 2.98 |
ENSDART00000115278
|
cx47.1
|
connexin 47.1 |
| chr5_+_29820266 | 2.93 |
ENSDART00000146331
ENSDART00000098315 |
f11r.2
|
F11 receptor, tandem duplicate 2 |
| chr9_+_21146862 | 2.91 |
ENSDART00000136365
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr7_-_52096498 | 2.89 |
ENSDART00000098688
ENSDART00000098690 |
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr14_+_36246726 | 2.76 |
ENSDART00000105602
|
elovl6
|
ELOVL fatty acid elongase 6 |
| chr18_+_30915348 | 2.75 |
ENSDART00000139398
|
mthfsd
|
methenyltetrahydrofolate synthetase domain containing |
| chr21_+_39963851 | 2.73 |
ENSDART00000144435
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr21_+_11385031 | 2.70 |
ENSDART00000040598
|
cel.2
|
carboxyl ester lipase, tandem duplicate 2 |
| chr25_-_1720736 | 2.65 |
ENSDART00000097256
|
SLC6A13
|
solute carrier family 6 member 13 |
| chr14_-_41285392 | 2.59 |
ENSDART00000147389
|
tmem35
|
transmembrane protein 35 |
| chr6_-_35472923 | 2.50 |
ENSDART00000185907
|
rgs8
|
regulator of G protein signaling 8 |
| chr8_-_45277370 | 2.48 |
ENSDART00000146364
|
adamts13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13 |
| chr4_-_14328997 | 2.43 |
ENSDART00000091151
|
nell2b
|
neural EGFL like 2b |
| chr15_-_33896159 | 2.25 |
ENSDART00000159791
|
mag
|
myelin associated glycoprotein |
| chr24_-_38079261 | 2.23 |
ENSDART00000105662
|
crp1
|
C-reactive protein 1 |
| chr9_-_16218097 | 2.20 |
ENSDART00000190503
|
myo1b
|
myosin IB |
| chr1_+_44800298 | 2.15 |
ENSDART00000073712
ENSDART00000187054 |
tmem187
|
transmembrane protein 187 |
| chr5_+_1278092 | 2.14 |
ENSDART00000147972
ENSDART00000159783 |
dnm1a
|
dynamin 1a |
| chr3_-_39190317 | 2.14 |
ENSDART00000013167
|
retsat
|
retinol saturase (all-trans-retinol 13,14-reductase) |
| chr13_+_18659325 | 2.09 |
ENSDART00000147905
ENSDART00000188249 ENSDART00000134740 |
selenou1a
|
selenoprotein U 1a |
| chr7_-_52096287 | 2.08 |
ENSDART00000174068
|
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr18_+_10926197 | 2.06 |
ENSDART00000192387
|
ttc38
|
tetratricopeptide repeat domain 38 |
| chr11_+_35364445 | 2.06 |
ENSDART00000125766
|
camkvb
|
CaM kinase-like vesicle-associated b |
| chr10_+_4235998 | 2.02 |
ENSDART00000169328
|
plppr1
|
phospholipid phosphatase related 1 |
| chr8_+_24747865 | 1.93 |
ENSDART00000078656
|
slc16a4
|
solute carrier family 16, member 4 |
| chr17_-_33289304 | 1.89 |
ENSDART00000135118
ENSDART00000040346 |
efr3ba
|
EFR3 homolog Ba (S. cerevisiae) |
| chr10_-_22845485 | 1.87 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr1_-_12278056 | 1.85 |
ENSDART00000139336
ENSDART00000137463 |
cplx2l
|
complexin 2, like |
| chr7_+_23495986 | 1.84 |
ENSDART00000190739
ENSDART00000115299 ENSDART00000101423 ENSDART00000142401 |
zgc:109889
|
zgc:109889 |
| chr7_-_40122139 | 1.76 |
ENSDART00000173982
|
si:ch73-174h16.5
|
si:ch73-174h16.5 |
| chr3_-_55525627 | 1.76 |
ENSDART00000189234
|
tex2
|
testis expressed 2 |
| chr16_+_43152727 | 1.75 |
ENSDART00000125590
ENSDART00000154493 |
adam22
|
ADAM metallopeptidase domain 22 |
| chr24_-_33276139 | 1.75 |
ENSDART00000128943
|
nrbp2b
|
nuclear receptor binding protein 2b |
| chr19_-_15397946 | 1.73 |
ENSDART00000143480
|
hivep3a
|
human immunodeficiency virus type I enhancer binding protein 3a |
| chr8_+_6967108 | 1.70 |
ENSDART00000004588
|
asic1a
|
acid-sensing (proton-gated) ion channel 1a |
| chr3_-_40976463 | 1.65 |
ENSDART00000128450
ENSDART00000018676 |
cyp3c1
|
cytochrome P450, family 3, subfamily c, polypeptide 1 |
| chr16_-_25563508 | 1.64 |
ENSDART00000189642
|
atxn1b
|
ataxin 1b |
| chr18_-_34549721 | 1.62 |
ENSDART00000137101
ENSDART00000021880 |
ssr3
|
signal sequence receptor, gamma |
| chr16_+_11623956 | 1.59 |
ENSDART00000137788
|
cxcr3.1
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 1 |
| chr14_-_3174570 | 1.57 |
ENSDART00000163585
|
csf1ra
|
colony stimulating factor 1 receptor, a |
| chr14_-_46113321 | 1.54 |
ENSDART00000169040
ENSDART00000161475 ENSDART00000124925 |
si:ch211-235e9.8
|
si:ch211-235e9.8 |
| chr14_-_9350364 | 1.52 |
ENSDART00000164451
|
fam46d
|
family with sequence similarity 46, member D |
| chr21_-_7178348 | 1.50 |
ENSDART00000187467
|
fam69b
|
family with sequence similarity 69, member B |
| chr18_+_30878196 | 1.50 |
ENSDART00000099326
ENSDART00000146041 |
mthfsd
|
methenyltetrahydrofolate synthetase domain containing |
| chr16_+_4138331 | 1.48 |
ENSDART00000192403
ENSDART00000193036 |
mtf1
|
metal-regulatory transcription factor 1 |
| chr1_-_42779075 | 1.47 |
ENSDART00000133917
|
lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr14_+_29609245 | 1.46 |
ENSDART00000043058
|
TENM2
|
si:dkey-34l15.2 |
| chr3_-_18711288 | 1.44 |
ENSDART00000183885
|
grid2ipa
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, a |
| chr7_-_18601206 | 1.44 |
ENSDART00000111636
|
DTX4
|
si:ch211-119e14.2 |
| chr1_-_44704261 | 1.42 |
ENSDART00000133210
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr13_+_11439486 | 1.42 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr10_-_2522588 | 1.41 |
ENSDART00000081926
|
CU856539.1
|
|
| chr4_-_50301491 | 1.41 |
ENSDART00000163146
|
si:ch211-197e7.3
|
si:ch211-197e7.3 |
| chr15_+_22267847 | 1.39 |
ENSDART00000110665
|
spa17
|
sperm autoantigenic protein 17 |
| chr24_-_33284945 | 1.38 |
ENSDART00000155429
ENSDART00000112845 |
zgc:195173
|
zgc:195173 |
| chr14_+_22447662 | 1.37 |
ENSDART00000161776
|
sowahab
|
sosondowah ankyrin repeat domain family member Ab |
| chr3_+_18795570 | 1.37 |
ENSDART00000042368
|
fahd1
|
fumarylacetoacetate hydrolase domain containing 1 |
| chr6_+_6780873 | 1.36 |
ENSDART00000011865
|
sec23b
|
Sec23 homolog B, COPII coat complex component |
| chr6_-_43092175 | 1.35 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr9_+_31534601 | 1.35 |
ENSDART00000133094
|
si:ch211-168k14.2
|
si:ch211-168k14.2 |
| chr9_+_19421841 | 1.34 |
ENSDART00000159090
ENSDART00000084771 |
pde9a
|
phosphodiesterase 9A |
| chr3_+_18097700 | 1.33 |
ENSDART00000021634
|
wfikkn2a
|
info WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2a |
| chr15_+_32423801 | 1.32 |
ENSDART00000165591
|
si:dkey-285b23.3
|
si:dkey-285b23.3 |
| chr14_-_30897177 | 1.32 |
ENSDART00000087918
|
slc7a3b
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3b |
| chr25_+_4760489 | 1.32 |
ENSDART00000167399
|
FP245455.1
|
|
| chr3_-_13068189 | 1.31 |
ENSDART00000167180
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr1_-_52498146 | 1.31 |
ENSDART00000122217
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr17_+_25289431 | 1.31 |
ENSDART00000161002
|
kbtbd11
|
kelch repeat and BTB (POZ) domain containing 11 |
| chr1_+_6817292 | 1.31 |
ENSDART00000145822
ENSDART00000092118 |
erbb4a
|
erb-b2 receptor tyrosine kinase 4a |
| chr7_+_26100024 | 1.29 |
ENSDART00000173726
|
si:ch211-196f2.3
|
si:ch211-196f2.3 |
| chr5_+_67812062 | 1.29 |
ENSDART00000158611
|
zgc:175280
|
zgc:175280 |
| chr12_-_31103906 | 1.28 |
ENSDART00000189099
ENSDART00000121527 ENSDART00000185635 ENSDART00000183754 |
tcf7l2
|
transcription factor 7 like 2 |
| chr17_-_52595932 | 1.26 |
ENSDART00000127225
|
si:ch211-173a9.7
|
si:ch211-173a9.7 |
| chr14_-_24101897 | 1.23 |
ENSDART00000143695
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr11_+_15931188 | 1.20 |
ENSDART00000165768
ENSDART00000081098 |
draxin
|
dorsal inhibitory axon guidance protein |
| chr18_+_17534627 | 1.20 |
ENSDART00000061007
|
mt2
|
metallothionein 2 |
| chr5_-_33606729 | 1.20 |
ENSDART00000137073
|
si:dkey-34e4.1
|
si:dkey-34e4.1 |
| chr9_+_16854121 | 1.20 |
ENSDART00000110866
|
cln5
|
CLN5, intracellular trafficking protein |
| chr5_-_54790923 | 1.19 |
ENSDART00000176835
|
pik3r1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr22_+_25274712 | 1.18 |
ENSDART00000137341
|
BX950205.1
|
|
| chr7_-_71384391 | 1.17 |
ENSDART00000112841
|
ccdc149a
|
coiled-coil domain containing 149a |
| chr22_+_12361317 | 1.17 |
ENSDART00000189963
ENSDART00000159614 |
r3hdm1
|
R3H domain containing 1 |
| chr1_+_53321878 | 1.16 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr17_-_52594756 | 1.15 |
ENSDART00000167288
|
si:ch211-173a9.7
|
si:ch211-173a9.7 |
| chr4_-_63980140 | 1.14 |
ENSDART00000160818
|
znf1091
|
zinc finger protein 1091 |
| chr22_+_12361489 | 1.14 |
ENSDART00000182483
|
r3hdm1
|
R3H domain containing 1 |
| chr17_-_20218357 | 1.11 |
ENSDART00000155990
ENSDART00000155632 ENSDART00000156540 ENSDART00000063523 |
mgmt
|
O-6-methylguanine-DNA methyltransferase |
| chr4_+_25693463 | 1.11 |
ENSDART00000132864
|
acot18
|
acyl-CoA thioesterase 18 |
| chr2_+_55199721 | 1.10 |
ENSDART00000016143
|
zmp:0000000521
|
zmp:0000000521 |
| chr14_+_29941445 | 1.09 |
ENSDART00000181761
|
fam149a
|
family with sequence similarity 149 member A |
| chr15_+_1765988 | 1.08 |
ENSDART00000159032
|
cul3b
|
cullin 3b |
| chr6_+_29923593 | 1.08 |
ENSDART00000169687
|
dlg1
|
discs, large homolog 1 (Drosophila) |
| chr3_-_40976288 | 1.08 |
ENSDART00000193553
|
cyp3c1
|
cytochrome P450, family 3, subfamily c, polypeptide 1 |
| chr4_+_9030609 | 1.07 |
ENSDART00000154399
|
aldh1l2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr18_-_15771551 | 1.07 |
ENSDART00000130931
ENSDART00000154079 |
si:ch211-219a15.3
|
si:ch211-219a15.3 |
| chr21_+_18353703 | 1.06 |
ENSDART00000181396
ENSDART00000166359 |
si:ch73-287m6.1
|
si:ch73-287m6.1 |
| chr21_-_2248239 | 1.06 |
ENSDART00000169448
|
si:ch73-299h12.1
|
si:ch73-299h12.1 |
| chr23_+_25201077 | 1.06 |
ENSDART00000136675
|
si:dkey-151g10.3
|
si:dkey-151g10.3 |
| chr23_-_45319592 | 1.06 |
ENSDART00000189861
|
ccdc171
|
coiled-coil domain containing 171 |
| chr17_+_24756379 | 1.05 |
ENSDART00000185059
|
stac
|
SH3 and cysteine rich domain |
| chr19_+_42955280 | 1.04 |
ENSDART00000141225
|
fabp3
|
fatty acid binding protein 3, muscle and heart |
| chr14_+_3411771 | 1.04 |
ENSDART00000164778
|
trpc3
|
transient receptor potential cation channel, subfamily C, member 3 |
| chr11_-_29833698 | 1.02 |
ENSDART00000079149
|
xk
|
X-linked Kx blood group (McLeod syndrome) |
| chr11_+_30636351 | 1.02 |
ENSDART00000087909
|
tmem246
|
transmembrane protein 246 |
| chr3_+_15279016 | 1.01 |
ENSDART00000182005
|
asphd1
|
aspartate beta-hydroxylase domain containing 1 |
| chr14_+_25464681 | 1.01 |
ENSDART00000067500
ENSDART00000187601 |
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr9_-_30346279 | 1.01 |
ENSDART00000089488
|
sytl5
|
synaptotagmin-like 5 |
| chr12_-_41662327 | 0.99 |
ENSDART00000191602
ENSDART00000170423 |
jakmip3
|
Janus kinase and microtubule interacting protein 3 |
| chr18_-_16181952 | 0.99 |
ENSDART00000157824
|
slc6a15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr20_+_25911342 | 0.99 |
ENSDART00000146004
|
ttbk2b
|
tau tubulin kinase 2b |
| chr11_-_27537593 | 0.98 |
ENSDART00000173444
ENSDART00000172895 ENSDART00000088177 |
ptpdc1a
|
protein tyrosine phosphatase domain containing 1a |
| chr15_-_33904831 | 0.98 |
ENSDART00000164333
ENSDART00000165404 |
mag
|
myelin associated glycoprotein |
| chr17_-_19963718 | 0.98 |
ENSDART00000154251
|
chrm3a
|
cholinergic receptor, muscarinic 3a |
| chr19_-_36234185 | 0.97 |
ENSDART00000186003
|
csmd2
|
CUB and Sushi multiple domains 2 |
| chr20_+_21583639 | 0.97 |
ENSDART00000131069
|
esr2a
|
estrogen receptor 2a |
| chr16_-_25400257 | 0.97 |
ENSDART00000040756
|
zgc:136493
|
zgc:136493 |
| chr21_+_33503835 | 0.96 |
ENSDART00000125658
|
clint1b
|
clathrin interactor 1b |
| chr2_-_51630555 | 0.96 |
ENSDART00000171746
|
pigr
|
polymeric immunoglobulin receptor |
| chr22_+_1779401 | 0.95 |
ENSDART00000170126
|
znf1154
|
zinc finger protein 1154 |
| chr16_+_50741154 | 0.94 |
ENSDART00000101627
|
IGLON5
|
zgc:110372 |
| chr2_-_42843797 | 0.94 |
ENSDART00000137913
|
adcy8
|
adenylate cyclase 8 (brain) |
| chr20_-_35040041 | 0.94 |
ENSDART00000131919
|
kif26bb
|
kinesin family member 26Bb |
| chr2_-_10062575 | 0.93 |
ENSDART00000091726
|
fam78ba
|
family with sequence similarity 78, member B a |
| chr1_-_49649122 | 0.93 |
ENSDART00000134399
|
slkb
|
STE20-like kinase b |
| chr16_+_5184402 | 0.91 |
ENSDART00000156685
|
soga3a
|
SOGA family member 3a |
| chr14_+_6954579 | 0.91 |
ENSDART00000060998
|
nme5
|
NME/NM23 family member 5 |
| chr7_+_33145529 | 0.90 |
ENSDART00000052387
|
itln2
|
intelectin 2 |
| chr11_-_41996957 | 0.89 |
ENSDART00000055706
|
her15.2
|
hairy and enhancer of split-related 15, tandem duplicate 2 |
| chr7_-_39378903 | 0.88 |
ENSDART00000173659
|
slc8b1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr20_+_40457599 | 0.87 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr4_-_75803810 | 0.87 |
ENSDART00000163517
|
si:ch211-203c5.3
|
si:ch211-203c5.3 |
| chr6_-_7842078 | 0.87 |
ENSDART00000065507
|
plppr2b
|
phospholipid phosphatase related 2b |
| chr14_+_17382803 | 0.86 |
ENSDART00000040383
|
si:ch211-255i20.3
|
si:ch211-255i20.3 |
| chr24_-_25673405 | 0.86 |
ENSDART00000186081
ENSDART00000110241 ENSDART00000142351 |
cnksr2a
|
connector enhancer of kinase suppressor of Ras 2a |
| chr21_-_37972412 | 0.85 |
ENSDART00000186315
|
ripply1
|
ripply transcriptional repressor 1 |
| chr2_-_12502495 | 0.85 |
ENSDART00000186781
|
dpp6b
|
dipeptidyl-peptidase 6b |
| chr11_+_6152643 | 0.84 |
ENSDART00000012789
|
slc1a6
|
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 |
| chr14_+_25817628 | 0.83 |
ENSDART00000047680
|
glra1
|
glycine receptor, alpha 1 |
| chr2_+_24867534 | 0.83 |
ENSDART00000158050
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr17_-_13070602 | 0.82 |
ENSDART00000188311
ENSDART00000193428 |
CU469462.1
|
|
| chr21_+_21791343 | 0.81 |
ENSDART00000151654
|
neu3.1
|
sialidase 3 (membrane sialidase), tandem duplicate 1 |
| chr8_+_13368150 | 0.80 |
ENSDART00000114699
|
slc5a5
|
solute carrier family 5 (sodium/iodide cotransporter), member 5 |
| chr11_-_1948784 | 0.80 |
ENSDART00000082475
|
nr1d4b
|
nuclear receptor subfamily 1, group D, member 4b |
| chr5_+_45990046 | 0.80 |
ENSDART00000084024
|
sv2c
|
synaptic vesicle glycoprotein 2C |
| chr12_+_2870671 | 0.80 |
ENSDART00000165225
|
prkar1ab
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) b |
| chr20_-_36227500 | 0.79 |
ENSDART00000166054
|
BX255920.1
|
|
| chr13_+_41917606 | 0.79 |
ENSDART00000114741
|
polr1b
|
polymerase (RNA) I polypeptide B |
| chr13_+_40034176 | 0.76 |
ENSDART00000189797
|
golga7bb
|
golgin A7 family, member Bb |
| chr10_-_3258073 | 0.75 |
ENSDART00000113162
|
pi4kaa
|
phosphatidylinositol 4-kinase, catalytic, alpha a |
| chr4_+_47445318 | 0.75 |
ENSDART00000183819
|
si:dkey-124l13.1
|
si:dkey-124l13.1 |
| chr1_-_39977551 | 0.74 |
ENSDART00000139354
|
stox2a
|
storkhead box 2a |
| chr17_-_6519423 | 0.74 |
ENSDART00000175626
|
CR628323.3
|
|
| chr7_-_35036770 | 0.74 |
ENSDART00000123174
|
galr1b
|
galanin receptor 1b |
| chr17_-_6399920 | 0.73 |
ENSDART00000022010
|
hivep2b
|
human immunodeficiency virus type I enhancer binding protein 2b |
| chr1_+_29281764 | 0.73 |
ENSDART00000112106
|
fam155a
|
family with sequence similarity 155, member A |
| chr6_+_6828167 | 0.72 |
ENSDART00000181284
|
si:ch211-85n16.4
|
si:ch211-85n16.4 |
| chr18_+_38807239 | 0.71 |
ENSDART00000184332
|
fam214a
|
family with sequence similarity 214, member A |
| chr8_-_43120844 | 0.71 |
ENSDART00000193242
|
ccdc92
|
coiled-coil domain containing 92 |
| chr25_+_22682065 | 0.70 |
ENSDART00000129484
|
ush1c
|
Usher syndrome 1C |
| chr23_-_15036789 | 0.69 |
ENSDART00000056570
|
phf20b
|
PHD finger protein 20, b |
| chr6_-_39198912 | 0.69 |
ENSDART00000077938
|
c1galt1b
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1b |
| chr13_-_40316367 | 0.69 |
ENSDART00000009343
|
pyroxd2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr7_-_34329527 | 0.69 |
ENSDART00000173454
|
madd
|
MAP-kinase activating death domain |
| chr17_+_18031899 | 0.68 |
ENSDART00000022758
|
setd3
|
SET domain containing 3 |
| chr16_+_25074029 | 0.68 |
ENSDART00000155465
|
si:dkeyp-84f3.9
|
si:dkeyp-84f3.9 |
| chr20_+_18945057 | 0.67 |
ENSDART00000035447
|
mtmr9
|
myotubularin related protein 9 |
| chr7_+_65876335 | 0.67 |
ENSDART00000150143
|
tead1b
|
TEA domain family member 1b |
| chr13_-_31687925 | 0.67 |
ENSDART00000085989
|
trmt5
|
TRM5 tRNA methyltransferase 5 homolog (S. cerevisiae) |
| chr21_-_1742159 | 0.66 |
ENSDART00000151049
|
onecut2
|
one cut homeobox 2 |
| chr1_-_57172294 | 0.66 |
ENSDART00000063774
|
rac1l
|
Rac family small GTPase 1, like |
| chr3_-_25119839 | 0.65 |
ENSDART00000154724
|
chadla
|
chondroadherin-like a |
| chr10_+_33627429 | 0.65 |
ENSDART00000138542
|
b3glctb
|
beta 3-glucosyltransferase b |
| chr2_-_9260002 | 0.65 |
ENSDART00000057299
|
st6galnac5a
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5a |
| chr5_-_41860556 | 0.65 |
ENSDART00000140154
|
si:dkey-65b12.12
|
si:dkey-65b12.12 |
| chr17_-_20218167 | 0.64 |
ENSDART00000154479
|
mgmt
|
O-6-methylguanine-DNA methyltransferase |
| chr6_+_54498220 | 0.64 |
ENSDART00000103282
|
si:ch211-233f11.5
|
si:ch211-233f11.5 |
| chr16_-_52736549 | 0.64 |
ENSDART00000146607
|
azin1a
|
antizyme inhibitor 1a |
| chr4_+_13412030 | 0.64 |
ENSDART00000003694
|
cand1
|
cullin-associated and neddylation-dissociated 1 |
| chr23_-_36670369 | 0.63 |
ENSDART00000006881
|
zbtb39
|
zinc finger and BTB domain containing 39 |
| chr12_+_29240124 | 0.63 |
ENSDART00000053761
ENSDART00000130172 |
bms1
|
BMS1 ribosome biogenesis factor |
| chr9_+_48085039 | 0.63 |
ENSDART00000186734
|
si:ch73-54b5.2
|
si:ch73-54b5.2 |
| chr21_-_12036134 | 0.63 |
ENSDART00000031658
|
tpgs2
|
tubulin polyglutamylase complex subunit 2 |
| chr14_+_11909966 | 0.62 |
ENSDART00000171829
|
frmpd3
|
FERM and PDZ domain containing 3 |
| chr21_-_37973081 | 0.61 |
ENSDART00000136569
|
ripply1
|
ripply transcriptional repressor 1 |
| chr23_-_44470253 | 0.61 |
ENSDART00000176333
|
si:ch1073-228j22.2
|
si:ch1073-228j22.2 |
| chr7_-_24204200 | 0.60 |
ENSDART00000087298
|
gmpr2
|
guanosine monophosphate reductase 2 |
| chr23_-_10048533 | 0.60 |
ENSDART00000166663
|
plxnb1a
|
plexin b1a |
| chr4_-_71904377 | 0.60 |
ENSDART00000191852
|
si:dkeyp-4f2.1
|
si:dkeyp-4f2.1 |
| chr15_-_30815826 | 0.60 |
ENSDART00000156160
ENSDART00000145918 |
msi2b
|
musashi RNA-binding protein 2b |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox17
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.1 | GO:0010873 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 1.2 | 3.7 | GO:0043606 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.9 | 7.0 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.6 | 2.5 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.4 | 1.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.4 | 2.6 | GO:0043091 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.4 | 3.2 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.4 | 1.6 | GO:0071674 | regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) regulation of odontogenesis(GO:0042481) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) |
| 0.4 | 1.5 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.4 | 1.1 | GO:0009258 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.3 | 1.0 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 0.3 | 1.3 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.3 | 0.9 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 0.3 | 0.8 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.3 | 0.8 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.2 | 1.0 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.2 | 2.7 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.2 | 1.7 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.2 | 0.9 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 0.6 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.2 | 2.9 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.2 | 1.0 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.2 | 0.8 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.2 | 1.2 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.2 | 1.4 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.1 | 2.1 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 2.8 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.9 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.4 | GO:0072047 | proximal/distal pattern formation involved in nephron development(GO:0072047) distal tubule morphogenesis(GO:0072156) proximal/distal pattern formation involved in pronephric nephron development(GO:0072196) |
| 0.1 | 1.8 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.4 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.6 | GO:1902047 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 1.2 | GO:0010043 | response to zinc ion(GO:0010043) |
| 0.1 | 3.4 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.1 | 0.4 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.1 | 1.0 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 1.6 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 1.2 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 1.5 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 4.7 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 1.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 1.5 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.1 | 0.7 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 1.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 1.4 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 0.2 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.1 | 0.4 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.1 | 1.4 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.1 | 1.4 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.7 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.1 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.4 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 0.8 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 0.9 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 1.1 | GO:1903286 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 0.9 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.1 | 0.2 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.1 | 0.7 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.1 | 0.8 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.1 | 0.3 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 1.6 | GO:0099633 | protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.1 | 4.7 | GO:0048741 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.1 | 1.1 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 0.2 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.1 | 0.1 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.1 | 1.1 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.2 | GO:1902592 | virion assembly(GO:0019068) virus maturation(GO:0019075) multi-organism membrane organization(GO:0044803) viral budding(GO:0046755) multi-organism organelle organization(GO:1902590) multi-organism membrane budding(GO:1902592) |
| 0.0 | 1.2 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.7 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.9 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.7 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.4 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.3 | GO:0035475 | angioblast cell migration involved in selective angioblast sprouting(GO:0035475) |
| 0.0 | 1.9 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.4 | GO:0071357 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.1 | GO:1901232 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.0 | 0.6 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.3 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.9 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 1.2 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.6 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.7 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.3 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 2.6 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.4 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 2.1 | GO:0007599 | hemostasis(GO:0007599) |
| 0.0 | 0.3 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.6 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.6 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 1.1 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 1.9 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 1.6 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.7 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.8 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.8 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.2 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.0 | 0.3 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.5 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.6 | GO:0010906 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.3 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 1.1 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.6 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 1.1 | GO:0009755 | hormone-mediated signaling pathway(GO:0009755) |
| 0.0 | 1.6 | GO:1990778 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.3 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 2.3 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.7 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 0.6 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 1.0 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 1.0 | GO:0098661 | inorganic anion transmembrane transport(GO:0098661) |
| 0.0 | 1.0 | GO:0016573 | histone acetylation(GO:0016573) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.3 | 1.3 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.2 | 2.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 0.6 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 2.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 0.5 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.1 | 2.2 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 0.4 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.1 | 1.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.4 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.7 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 1.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.7 | GO:0002139 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.1 | 1.4 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 1.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 0.6 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 0.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 1.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 2.6 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 1.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.8 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 1.9 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.7 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 1.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.3 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.6 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 3.3 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.4 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 1.0 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.6 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.8 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.6 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.4 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 1.4 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.9 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.5 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 1.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 1.0 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 2.3 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 1.2 | 6.1 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 1.1 | 3.4 | GO:0015562 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 1.0 | 3.0 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.9 | 2.7 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.4 | 2.6 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.4 | 1.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.3 | 1.7 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.3 | 1.0 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.3 | 2.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.3 | 0.8 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.3 | 1.4 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.2 | 0.6 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.2 | 1.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.2 | 1.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 1.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.2 | 1.4 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.2 | 3.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 1.8 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.2 | 2.7 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.2 | 0.6 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 1.0 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.1 | 1.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 2.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.7 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 2.9 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 2.8 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.7 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 3.9 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 1.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 7.5 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 0.4 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 1.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.8 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.6 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 1.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 3.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.8 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.7 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 1.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.2 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.1 | 1.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.8 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 1.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.4 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 0.6 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 1.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.4 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 1.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 3.4 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 3.7 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 1.1 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.5 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 2.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.9 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.4 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.3 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 6.9 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.9 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.6 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 1.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 1.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 0.2 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.9 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.4 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.9 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 1.9 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 2.1 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.8 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 2.4 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 0.4 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.7 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.8 | GO:0015370 | solute:sodium symporter activity(GO:0015370) |
| 0.0 | 0.4 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 3.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.3 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 1.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.8 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.6 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.4 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 1.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.9 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 1.3 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.2 | GO:0015293 | symporter activity(GO:0015293) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 1.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 1.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 0.4 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 0.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 1.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.0 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.1 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.0 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 4.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.3 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.3 | 2.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.3 | 3.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.3 | 1.0 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.2 | 0.8 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 2.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 2.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 2.3 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 1.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.6 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.1 | 1.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 0.8 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 0.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.8 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 2.5 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 2.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.4 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |