Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
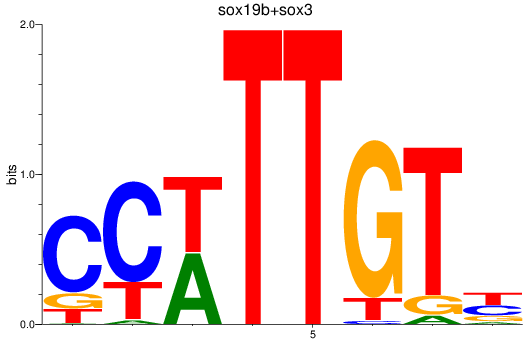
Results for sox19b+sox3
Z-value: 1.02
Transcription factors associated with sox19b+sox3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox19b
|
ENSDARG00000040266 | SRY-box transcription factor 19b |
|
sox3
|
ENSDARG00000053569 | SRY-box transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox3 | dr11_v1_chr14_-_32744464_32744464 | 0.32 | 1.5e-03 | Click! |
| sox19b | dr11_v1_chr7_-_26497947_26497947 | 0.28 | 6.5e-03 | Click! |
Activity profile of sox19b+sox3 motif
Sorted Z-values of sox19b+sox3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_21801277 | 11.74 |
ENSDART00000088407
|
trim108
|
tripartite motif containing 108 |
| chr5_-_30615901 | 8.06 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr13_+_22675802 | 8.05 |
ENSDART00000145538
ENSDART00000143312 |
zgc:193505
|
zgc:193505 |
| chr10_+_33171501 | 7.49 |
ENSDART00000159666
|
myl10
|
myosin, light chain 10, regulatory |
| chr14_+_38786298 | 7.15 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr23_-_10175898 | 7.01 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr21_+_25688388 | 6.68 |
ENSDART00000125709
|
bicdl2
|
bicaudal-D-related protein 2 |
| chr7_+_35268880 | 6.65 |
ENSDART00000182231
|
dpep2
|
dipeptidase 2 |
| chr14_-_8080416 | 6.59 |
ENSDART00000045109
|
zgc:92242
|
zgc:92242 |
| chr5_-_55560937 | 6.45 |
ENSDART00000148436
|
gna14
|
guanine nucleotide binding protein (G protein), alpha 14 |
| chr5_+_23256187 | 6.01 |
ENSDART00000168717
ENSDART00000142915 |
si:dkey-125i10.3
|
si:dkey-125i10.3 |
| chr3_-_57425961 | 5.90 |
ENSDART00000033716
|
socs3a
|
suppressor of cytokine signaling 3a |
| chr25_+_35502552 | 5.82 |
ENSDART00000189612
ENSDART00000058443 |
fibina
|
fin bud initiation factor a |
| chr3_+_12755535 | 5.81 |
ENSDART00000161286
|
cyp2k17
|
cytochrome P450, family 2, subfamily K, polypeptide17 |
| chr16_+_23960744 | 5.74 |
ENSDART00000058965
|
apoeb
|
apolipoprotein Eb |
| chr19_+_10937932 | 5.73 |
ENSDART00000168968
|
si:ch73-347e22.8
|
si:ch73-347e22.8 |
| chr3_-_50139860 | 5.73 |
ENSDART00000101563
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr14_+_11457500 | 5.71 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr12_+_48340133 | 5.57 |
ENSDART00000152899
ENSDART00000153335 ENSDART00000054788 |
ddit4
|
DNA-damage-inducible transcript 4 |
| chr23_+_42813415 | 5.55 |
ENSDART00000055577
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr7_-_7398350 | 5.55 |
ENSDART00000012637
|
zgc:101810
|
zgc:101810 |
| chr4_-_16354292 | 5.54 |
ENSDART00000139919
|
lum
|
lumican |
| chr16_+_23960933 | 5.53 |
ENSDART00000146077
|
apoeb
|
apolipoprotein Eb |
| chr24_+_26006730 | 5.41 |
ENSDART00000140384
ENSDART00000139184 |
ccl20b
|
chemokine (C-C motif) ligand 20b |
| chr5_-_46505691 | 5.33 |
ENSDART00000111589
ENSDART00000122966 ENSDART00000166907 |
hapln1a
|
hyaluronan and proteoglycan link protein 1a |
| chr3_-_31258459 | 5.26 |
ENSDART00000177928
|
LO017965.1
|
|
| chr3_+_3545825 | 5.12 |
ENSDART00000109060
|
CR589947.1
|
|
| chr8_-_31107537 | 5.11 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| chr17_-_8862424 | 4.99 |
ENSDART00000064633
|
nkl.4
|
NK-lysin tandem duplicate 4 |
| chr18_-_15467446 | 4.89 |
ENSDART00000187847
|
endouc
|
endonuclease, polyU-specific C |
| chr12_-_30523216 | 4.85 |
ENSDART00000152896
ENSDART00000153191 |
plekhs1
|
pleckstrin homology domain containing, family S member 1 |
| chr4_+_18843015 | 4.84 |
ENSDART00000152086
ENSDART00000066977 ENSDART00000132567 |
bik
|
BCL2 interacting killer |
| chr7_+_48761875 | 4.79 |
ENSDART00000003690
|
acana
|
aggrecan a |
| chr16_-_9675982 | 4.79 |
ENSDART00000113724
|
mal2
|
mal, T cell differentiation protein 2 (gene/pseudogene) |
| chr24_-_11076400 | 4.77 |
ENSDART00000003195
|
chmp4c
|
charged multivesicular body protein 4C |
| chr12_-_30558694 | 4.76 |
ENSDART00000153417
|
si:ch211-28p3.3
|
si:ch211-28p3.3 |
| chr11_+_10548171 | 4.66 |
ENSDART00000191497
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr16_+_29586468 | 4.61 |
ENSDART00000148926
|
mcl1b
|
MCL1, BCL2 family apoptosis regulator b |
| chr7_+_22688781 | 4.60 |
ENSDART00000173509
|
ugt5g1
|
UDP glucuronosyltransferase 5 family, polypeptide G1 |
| chr15_+_37331585 | 4.58 |
ENSDART00000170715
|
zgc:171592
|
zgc:171592 |
| chr7_-_53117131 | 4.56 |
ENSDART00000169211
ENSDART00000168890 ENSDART00000172179 ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr24_+_25069609 | 4.53 |
ENSDART00000115165
|
amer2
|
APC membrane recruitment protein 2 |
| chr12_-_30540699 | 4.49 |
ENSDART00000167712
ENSDART00000102464 |
zgc:153920
|
zgc:153920 |
| chr22_-_10121880 | 4.44 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr22_+_19220459 | 4.38 |
ENSDART00000163070
|
si:dkey-21e2.7
|
si:dkey-21e2.7 |
| chr25_-_22187397 | 4.32 |
ENSDART00000123211
ENSDART00000139110 |
pkp3a
|
plakophilin 3a |
| chr5_+_15203421 | 4.29 |
ENSDART00000040826
|
tbx1
|
T-box 1 |
| chr16_-_31976269 | 4.28 |
ENSDART00000139664
|
styk1
|
serine/threonine/tyrosine kinase 1 |
| chr23_+_20110086 | 4.27 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr7_+_20019125 | 4.26 |
ENSDART00000186391
|
bcl6b
|
B-cell CLL/lymphoma 6, member B |
| chr1_+_31054915 | 4.18 |
ENSDART00000148968
|
itga6b
|
integrin, alpha 6b |
| chr19_+_24488403 | 4.12 |
ENSDART00000052421
|
txnipa
|
thioredoxin interacting protein a |
| chr5_+_54938634 | 4.09 |
ENSDART00000163050
ENSDART00000145765 |
cd180
|
CD180 molecule |
| chr5_-_30984010 | 4.08 |
ENSDART00000182367
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr21_+_33459524 | 4.05 |
ENSDART00000053205
|
cd74b
|
CD74 molecule, major histocompatibility complex, class II invariant chain b |
| chr19_-_10771558 | 4.03 |
ENSDART00000085165
|
slc9a3.2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3, tandem duplicate 2 |
| chr5_-_37117778 | 4.03 |
ENSDART00000149138
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr7_+_48761646 | 4.02 |
ENSDART00000017467
|
acana
|
aggrecan a |
| chr1_+_22654875 | 3.93 |
ENSDART00000019698
ENSDART00000161874 |
anxa5b
|
annexin A5b |
| chr1_-_18615063 | 3.91 |
ENSDART00000014916
|
klf3
|
Kruppel-like factor 3 (basic) |
| chr22_+_22010128 | 3.90 |
ENSDART00000172610
|
gna15.2
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 2 |
| chr24_+_20373605 | 3.75 |
ENSDART00000112349
|
plcd1a
|
phospholipase C, delta 1a |
| chr22_-_10541372 | 3.74 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr14_-_34059681 | 3.71 |
ENSDART00000003993
|
itk
|
IL2 inducible T cell kinase |
| chr4_+_4232562 | 3.70 |
ENSDART00000177529
|
smkr1
|
small lysine rich protein 1 |
| chr6_-_1223551 | 3.70 |
ENSDART00000182112
|
CABZ01063034.1
|
|
| chr6_+_12968101 | 3.60 |
ENSDART00000013781
|
mcm6
|
minichromosome maintenance complex component 6 |
| chr22_-_3914162 | 3.60 |
ENSDART00000187174
ENSDART00000190612 ENSDART00000187928 ENSDART00000057224 ENSDART00000184758 |
mhc1uma
|
major histocompatibility complex class I UMA |
| chr19_+_19762183 | 3.60 |
ENSDART00000163611
ENSDART00000187604 |
hoxa3a
|
homeobox A3a |
| chr22_+_30631072 | 3.59 |
ENSDART00000059970
|
zmp:0000000606
|
zmp:0000000606 |
| chr6_+_30668098 | 3.55 |
ENSDART00000112294
|
ttc22
|
tetratricopeptide repeat domain 22 |
| chr5_+_1589458 | 3.49 |
ENSDART00000167473
|
CABZ01076669.1
|
|
| chr9_+_23770666 | 3.45 |
ENSDART00000182493
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr6_-_52675630 | 3.40 |
ENSDART00000083830
|
sdc4
|
syndecan 4 |
| chr8_+_34731982 | 3.39 |
ENSDART00000066050
|
hpdb
|
4-hydroxyphenylpyruvate dioxygenase b |
| chr25_+_14109626 | 3.39 |
ENSDART00000109883
|
actc1c
|
actin, alpha, cardiac muscle 1c |
| chr7_+_38380135 | 3.30 |
ENSDART00000174005
|
rhpn2
|
rhophilin, Rho GTPase binding protein 2 |
| chr9_-_6380653 | 3.26 |
ENSDART00000078523
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr20_+_6533260 | 3.25 |
ENSDART00000135005
ENSDART00000166356 |
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr22_+_661711 | 3.22 |
ENSDART00000113795
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr5_-_41841892 | 3.21 |
ENSDART00000167089
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr23_+_42819221 | 3.20 |
ENSDART00000180495
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr10_-_25628555 | 3.20 |
ENSDART00000143978
|
tiam1a
|
T cell lymphoma invasion and metastasis 1a |
| chr13_-_13294847 | 3.19 |
ENSDART00000125883
ENSDART00000013534 |
fgfr3
|
fibroblast growth factor receptor 3 |
| chr3_+_32125452 | 3.16 |
ENSDART00000110396
|
zgc:194125
|
zgc:194125 |
| chr23_-_14830627 | 3.12 |
ENSDART00000134659
|
sla2
|
Src-like-adaptor 2 |
| chr7_+_39410180 | 3.09 |
ENSDART00000168641
|
CT030188.1
|
|
| chr8_-_13013123 | 3.08 |
ENSDART00000147802
|
dennd2da
|
DENN/MADD domain containing 2Da |
| chr17_-_5860222 | 3.06 |
ENSDART00000058894
|
si:ch73-340m8.2
|
si:ch73-340m8.2 |
| chr22_+_18886209 | 3.05 |
ENSDART00000144402
|
fstl3
|
follistatin-like 3 (secreted glycoprotein) |
| chr8_-_39903803 | 3.05 |
ENSDART00000012391
|
cabp1a
|
calcium binding protein 1a |
| chr9_-_14084743 | 3.04 |
ENSDART00000056105
|
fer1l6
|
fer-1-like family member 6 |
| chr10_+_8690936 | 3.03 |
ENSDART00000144218
|
si:dkey-27b3.4
|
si:dkey-27b3.4 |
| chr14_+_10461264 | 3.02 |
ENSDART00000081095
|
cysltr1
|
cysteinyl leukotriene receptor 1 |
| chr8_-_36327328 | 3.01 |
ENSDART00000183333
|
zgc:103700
|
zgc:103700 |
| chr18_-_37241080 | 2.99 |
ENSDART00000126421
ENSDART00000078064 |
six9
|
SIX homeobox 9 |
| chr16_+_50089417 | 2.99 |
ENSDART00000153675
|
nr1d2a
|
nuclear receptor subfamily 1, group D, member 2a |
| chr22_+_3914318 | 2.98 |
ENSDART00000188774
ENSDART00000082034 |
FO904903.1
|
Danio rerio major histocompatibility complex class I ULA (mhc1ula), mRNA. |
| chr3_+_3573696 | 2.97 |
ENSDART00000169680
|
CR589947.2
|
|
| chr17_+_4408809 | 2.93 |
ENSDART00000113401
|
si:zfos-364h11.1
|
si:zfos-364h11.1 |
| chr22_+_19264181 | 2.92 |
ENSDART00000192750
ENSDART00000137591 ENSDART00000136197 |
si:dkey-78l4.5
si:dkey-21e2.11
|
si:dkey-78l4.5 si:dkey-21e2.11 |
| chr8_+_44358443 | 2.91 |
ENSDART00000189130
ENSDART00000189212 |
CU914776.2
|
|
| chr6_+_2030703 | 2.89 |
ENSDART00000109679
ENSDART00000187502 ENSDART00000191165 ENSDART00000187544 |
quo
|
quattro |
| chr9_-_42696408 | 2.89 |
ENSDART00000144744
|
col5a2a
|
collagen, type V, alpha 2a |
| chr3_-_32170850 | 2.87 |
ENSDART00000055307
ENSDART00000157366 |
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr16_+_21790870 | 2.84 |
ENSDART00000155039
|
trim108
|
tripartite motif containing 108 |
| chr7_+_29293452 | 2.83 |
ENSDART00000127358
|
si:ch211-112g6.4
|
si:ch211-112g6.4 |
| chr14_+_21222287 | 2.83 |
ENSDART00000159905
|
si:ch211-175m2.4
|
si:ch211-175m2.4 |
| chr15_-_36533322 | 2.83 |
ENSDART00000156466
ENSDART00000121755 |
si:dkey-262k9.4
|
si:dkey-262k9.4 |
| chr1_-_40735381 | 2.81 |
ENSDART00000093269
|
zgc:153642
|
zgc:153642 |
| chr20_+_38257766 | 2.75 |
ENSDART00000147485
ENSDART00000149160 |
ccl38.6
|
chemokine (C-C motif) ligand 38, duplicate 6 |
| chr23_-_28141419 | 2.74 |
ENSDART00000133039
|
tac3a
|
tachykinin 3a |
| chr1_+_44906517 | 2.73 |
ENSDART00000142820
|
wu:fc21g02
|
wu:fc21g02 |
| chr23_-_5683147 | 2.70 |
ENSDART00000102766
ENSDART00000067351 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr19_+_12915498 | 2.70 |
ENSDART00000132892
|
cthrc1a
|
collagen triple helix repeat containing 1a |
| chr12_-_44010532 | 2.64 |
ENSDART00000183875
|
si:ch211-182p11.1
|
si:ch211-182p11.1 |
| chr3_+_57820913 | 2.62 |
ENSDART00000168101
|
CU571328.1
|
|
| chr24_-_19719240 | 2.61 |
ENSDART00000135405
|
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr7_+_2245815 | 2.60 |
ENSDART00000114434
|
si:ch211-285c6.4
|
si:ch211-285c6.4 |
| chr12_-_36268723 | 2.60 |
ENSDART00000113740
|
kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr1_-_26027327 | 2.58 |
ENSDART00000171292
ENSDART00000170878 |
si:ch211-145b13.6
|
si:ch211-145b13.6 |
| chr8_-_46509194 | 2.57 |
ENSDART00000038924
|
sult1st1
|
sulfotransferase family 1, cytosolic sulfotransferase 1 |
| chr1_+_218524 | 2.56 |
ENSDART00000109529
|
tmco3
|
transmembrane and coiled-coil domains 3 |
| chr22_+_15898221 | 2.56 |
ENSDART00000062587
|
klf2a
|
Kruppel-like factor 2a |
| chr7_-_51368681 | 2.55 |
ENSDART00000146385
|
arhgap36
|
Rho GTPase activating protein 36 |
| chr21_+_11776328 | 2.55 |
ENSDART00000134469
ENSDART00000081646 |
glrx
|
glutaredoxin (thioltransferase) |
| chr21_+_27189490 | 2.55 |
ENSDART00000125349
|
bada
|
BCL2 associated agonist of cell death a |
| chr18_-_7481036 | 2.52 |
ENSDART00000101292
|
si:dkey-238c7.16
|
si:dkey-238c7.16 |
| chr7_+_39410393 | 2.51 |
ENSDART00000158561
ENSDART00000185173 |
CT030188.1
|
|
| chr23_+_35918530 | 2.49 |
ENSDART00000049551
|
rarga
|
retinoic acid receptor gamma a |
| chr9_-_24244383 | 2.48 |
ENSDART00000182407
|
cavin2a
|
caveolae associated protein 2a |
| chr12_-_30777540 | 2.45 |
ENSDART00000126466
|
entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr15_+_12429206 | 2.45 |
ENSDART00000168997
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr18_-_6634424 | 2.44 |
ENSDART00000062423
ENSDART00000179955 |
tnni1c
|
troponin I, skeletal, slow c |
| chr15_+_22448456 | 2.44 |
ENSDART00000040542
ENSDART00000190270 ENSDART00000185897 |
arhgef12a
|
Rho guanine nucleotide exchange factor (GEF) 12a |
| chr5_+_27898226 | 2.44 |
ENSDART00000098604
ENSDART00000180251 |
adam28
|
ADAM metallopeptidase domain 28 |
| chr2_+_2967255 | 2.43 |
ENSDART00000167649
ENSDART00000166449 |
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr17_+_45405821 | 2.42 |
ENSDART00000189482
ENSDART00000177422 |
tagapa
|
T cell activation RhoGTPase activating protein a |
| chr1_+_36651059 | 2.42 |
ENSDART00000187475
|
ednraa
|
endothelin receptor type Aa |
| chr6_+_54576520 | 2.42 |
ENSDART00000093199
ENSDART00000127519 ENSDART00000157142 |
tead3b
|
TEA domain family member 3 b |
| chr3_-_32956808 | 2.42 |
ENSDART00000183902
|
casp6l1
|
caspase 6, apoptosis-related cysteine peptidase, like 1 |
| chr18_-_6633984 | 2.41 |
ENSDART00000185241
|
tnni1c
|
troponin I, skeletal, slow c |
| chr12_-_30777743 | 2.39 |
ENSDART00000148888
|
entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr20_+_34770197 | 2.39 |
ENSDART00000018304
|
mcm3
|
minichromosome maintenance complex component 3 |
| chr6_-_6993046 | 2.38 |
ENSDART00000053304
|
si:ch211-114n24.6
|
si:ch211-114n24.6 |
| chr5_+_4564233 | 2.38 |
ENSDART00000193435
|
CABZ01058647.1
|
|
| chr21_-_26495700 | 2.38 |
ENSDART00000109379
|
cd248b
|
CD248 molecule, endosialin b |
| chr6_-_40098641 | 2.37 |
ENSDART00000017402
|
ip6k2b
|
inositol hexakisphosphate kinase 2b |
| chr19_+_10603405 | 2.36 |
ENSDART00000151135
|
si:dkey-211g8.8
|
si:dkey-211g8.8 |
| chr3_+_33761549 | 2.36 |
ENSDART00000169337
|
ier2a
|
immediate early response 2a |
| chr2_-_23390779 | 2.36 |
ENSDART00000020136
|
ivns1abpb
|
influenza virus NS1A binding protein b |
| chr5_+_38680247 | 2.34 |
ENSDART00000132763
|
si:dkey-58f10.11
|
si:dkey-58f10.11 |
| chr9_-_30220818 | 2.34 |
ENSDART00000140929
|
si:dkey-100n23.3
|
si:dkey-100n23.3 |
| chr25_-_14433503 | 2.32 |
ENSDART00000103957
|
exoc3l1
|
exocyst complex component 3-like 1 |
| chr22_-_15693085 | 2.31 |
ENSDART00000141861
|
si:ch1073-396h14.1
|
si:ch1073-396h14.1 |
| chr9_-_7683799 | 2.26 |
ENSDART00000102713
|
si:ch73-199e17.1
|
si:ch73-199e17.1 |
| chr7_-_28681724 | 2.21 |
ENSDART00000162400
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr10_+_44042033 | 2.21 |
ENSDART00000190006
ENSDART00000046172 |
cryba4
|
crystallin, beta A4 |
| chr11_-_11890001 | 2.19 |
ENSDART00000081544
|
wipf2a
|
WAS/WASL interacting protein family, member 2a |
| chr23_+_45025909 | 2.19 |
ENSDART00000188105
|
hmgb2b
|
high mobility group box 2b |
| chr5_-_36949476 | 2.18 |
ENSDART00000047269
|
h3f3c
|
H3 histone, family 3C |
| chr16_-_21799407 | 2.18 |
ENSDART00000123717
|
FP015862.4
|
|
| chr15_+_22014029 | 2.17 |
ENSDART00000079504
|
ankk1
|
ankyrin repeat and kinase domain containing 1 |
| chr23_+_24085531 | 2.17 |
ENSDART00000139710
|
ttll10
|
tubulin tyrosine ligase-like family, member 10 |
| chr4_-_4261673 | 2.16 |
ENSDART00000150694
|
cd9b
|
CD9 molecule b |
| chr16_+_30316177 | 2.15 |
ENSDART00000109505
|
tmem79b
|
transmembrane protein 79b |
| chr10_+_24690534 | 2.14 |
ENSDART00000079549
|
tpte
|
transmembrane phosphatase with tensin homology |
| chr19_+_20778011 | 2.14 |
ENSDART00000024208
|
nutf2l
|
nuclear transport factor 2, like |
| chr2_-_27900518 | 2.13 |
ENSDART00000109561
ENSDART00000077720 |
zgc:163121
|
zgc:163121 |
| chr12_-_28848015 | 2.12 |
ENSDART00000153200
|
si:ch211-194k22.8
|
si:ch211-194k22.8 |
| chr11_+_8129536 | 2.12 |
ENSDART00000158112
ENSDART00000011183 |
prkacba
|
protein kinase, cAMP-dependent, catalytic, beta a |
| chr3_-_16110351 | 2.12 |
ENSDART00000064838
|
lasp1
|
LIM and SH3 protein 1 |
| chr16_-_32208728 | 2.12 |
ENSDART00000023995
|
calhm5.2
|
calcium homeostasis modulator family member 5, tandem duplicate 2 |
| chr23_+_26079467 | 2.11 |
ENSDART00000129617
|
atp6ap1b
|
ATPase H+ transporting accessory protein 1b |
| chr18_+_40525408 | 2.09 |
ENSDART00000132833
|
vwa7
|
von Willebrand factor A domain containing 7 |
| chr21_+_22738939 | 2.08 |
ENSDART00000151342
ENSDART00000079145 |
arhgap42a
|
Rho GTPase activating protein 42a |
| chr10_+_29840725 | 2.08 |
ENSDART00000127268
|
BX120005.1
|
|
| chr11_+_27274355 | 2.08 |
ENSDART00000113707
|
fbln2
|
fibulin 2 |
| chr18_-_12327426 | 2.07 |
ENSDART00000136992
ENSDART00000114024 |
fam107b
|
family with sequence similarity 107, member B |
| chr18_+_48428713 | 2.06 |
ENSDART00000076861
|
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr19_+_47394270 | 2.06 |
ENSDART00000171281
|
psmb2
|
proteasome subunit beta 2 |
| chr23_+_42336084 | 2.04 |
ENSDART00000158959
ENSDART00000161812 |
cyp2aa7
cyp2aa8
|
cytochrome P450, family 2, subfamily AA, polypeptide 7 cytochrome P450, family 2, subfamily AA, polypeptide 8 |
| chr22_-_15577060 | 2.03 |
ENSDART00000176291
|
hsh2d
|
hematopoietic SH2 domain containing |
| chr1_-_49498116 | 2.02 |
ENSDART00000137357
|
zgc:175214
|
zgc:175214 |
| chr12_-_16990896 | 2.02 |
ENSDART00000152402
|
ifit12
|
interferon-induced protein with tetratricopeptide repeats 12 |
| chr18_+_44649804 | 2.02 |
ENSDART00000059063
|
ehd2b
|
EH-domain containing 2b |
| chr16_-_31598771 | 2.02 |
ENSDART00000016386
|
tg
|
thyroglobulin |
| chr1_-_8651718 | 2.01 |
ENSDART00000133319
|
actb1
|
actin, beta 1 |
| chr6_+_45932276 | 2.00 |
ENSDART00000103491
|
rbp7b
|
retinol binding protein 7b, cellular |
| chr23_+_25428513 | 2.00 |
ENSDART00000144554
|
fmnl3
|
formin-like 3 |
| chr19_+_4038589 | 2.00 |
ENSDART00000169271
|
btr23
|
bloodthirsty-related gene family, member 23 |
| chr4_-_63848305 | 1.99 |
ENSDART00000097308
|
BX901974.1
|
|
| chr15_-_20400423 | 1.99 |
ENSDART00000081290
ENSDART00000156813 ENSDART00000192427 |
rnaset2l
|
ribonuclease T2, like |
| chr5_-_51756210 | 1.98 |
ENSDART00000163464
|
lhfpl2b
|
LHFPL tetraspan subfamily member 2b |
| chr5_-_24127310 | 1.98 |
ENSDART00000182700
ENSDART00000154313 |
capga
|
capping protein (actin filament), gelsolin-like a |
| chr19_-_8940068 | 1.95 |
ENSDART00000043507
|
ciarta
|
circadian associated repressor of transcription a |
| chr15_-_25527580 | 1.95 |
ENSDART00000167005
ENSDART00000157498 |
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr24_-_40668208 | 1.94 |
ENSDART00000171543
|
smyhc1
|
slow myosin heavy chain 1 |
| chr11_+_29965822 | 1.94 |
ENSDART00000127049
|
il1fma
|
interleukin-1 family member A |
| chr22_-_10570749 | 1.91 |
ENSDART00000140736
|
si:dkey-42i9.6
|
si:dkey-42i9.6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox19b+sox3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.3 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 1.7 | 6.7 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 1.5 | 8.8 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 1.4 | 4.3 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 1.4 | 4.1 | GO:0060907 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 1.1 | 3.3 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 1.0 | 7.8 | GO:0070571 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.9 | 4.6 | GO:0045056 | transcytosis(GO:0045056) |
| 0.9 | 2.6 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.8 | 3.4 | GO:0048103 | neuronal stem cell division(GO:0036445) somatic stem cell division(GO:0048103) |
| 0.8 | 4.8 | GO:0071405 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.8 | 4.6 | GO:0090134 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.8 | 1.5 | GO:0061082 | positive regulation of myeloid leukocyte cytokine production involved in immune response(GO:0061081) myeloid leukocyte cytokine production(GO:0061082) |
| 0.7 | 4.4 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.7 | 4.8 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.7 | 3.4 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.6 | 1.9 | GO:0072196 | proximal/distal pattern formation involved in nephron development(GO:0072047) proximal/distal pattern formation involved in pronephric nephron development(GO:0072196) |
| 0.6 | 3.7 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.6 | 3.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.6 | 1.7 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.5 | 2.7 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.5 | 6.0 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.5 | 2.1 | GO:2000815 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.5 | 2.1 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.5 | 3.6 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.5 | 4.0 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.5 | 1.8 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.5 | 1.8 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.4 | 3.1 | GO:1901031 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.4 | 2.2 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.4 | 2.2 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.4 | 1.2 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.4 | 2.4 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.4 | 1.6 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.4 | 3.9 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.4 | 5.8 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.4 | 1.8 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.3 | 0.6 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.3 | 2.7 | GO:0042987 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.3 | 0.9 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.3 | 1.5 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.3 | 6.4 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.3 | 0.9 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.3 | 1.1 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.3 | 2.8 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.3 | 1.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.3 | 1.6 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.3 | 2.5 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.2 | 0.7 | GO:1903060 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.2 | 5.3 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.2 | 1.2 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.2 | 10.6 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 2.6 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.2 | 2.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 3.6 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.2 | 0.7 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.2 | 2.2 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.2 | 2.4 | GO:1900052 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.2 | 0.9 | GO:0090579 | establishment of mitotic sister chromatid cohesion(GO:0034087) establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.2 | 2.7 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.2 | 0.6 | GO:0071042 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.2 | 2.6 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.2 | 4.0 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.2 | 0.8 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.2 | 2.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.2 | 1.9 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.2 | 0.5 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.2 | 1.9 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.2 | 3.0 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 1.5 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.2 | 0.8 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.2 | 7.4 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.2 | 1.8 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.2 | 1.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 8.2 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.2 | 1.4 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.2 | 2.5 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.2 | 1.5 | GO:2000144 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 3.9 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.1 | 0.9 | GO:0055016 | hypochord development(GO:0055016) |
| 0.1 | 1.3 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 5.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 6.3 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 2.6 | GO:0006584 | catecholamine metabolic process(GO:0006584) catechol-containing compound metabolic process(GO:0009712) |
| 0.1 | 1.4 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 1.9 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 1.6 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.1 | 0.7 | GO:2000562 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.1 | 5.5 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 0.8 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 3.9 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 3.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 12.3 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.1 | 1.7 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.6 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.1 | 2.8 | GO:0003171 | atrioventricular valve development(GO:0003171) |
| 0.1 | 1.8 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.1 | 1.0 | GO:0042664 | negative regulation of endodermal cell fate specification(GO:0042664) |
| 0.1 | 1.2 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 3.1 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 0.7 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 0.5 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.1 | 1.4 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 2.4 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.1 | 0.4 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.1 | 1.3 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.1 | 1.3 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 4.8 | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043280) |
| 0.1 | 1.0 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.1 | 1.5 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.1 | 1.7 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.1 | 1.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 3.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 0.8 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.1 | 5.1 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.1 | 3.0 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 1.4 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.1 | 0.5 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.1 | 2.8 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.5 | GO:0003342 | bulbus arteriosus development(GO:0003232) proepicardium development(GO:0003342) septum transversum development(GO:0003343) |
| 0.1 | 2.2 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.1 | 1.9 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 6.6 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 0.3 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 1.0 | GO:0021654 | rhombomere boundary formation(GO:0021654) |
| 0.1 | 1.8 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.8 | GO:0031274 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 2.9 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 2.0 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 2.5 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 0.6 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.4 | GO:1902765 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.1 | 0.6 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 1.0 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.5 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 1.6 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.1 | 2.7 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.1 | 4.3 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.1 | 0.8 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 1.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.2 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.1 | 1.5 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 1.0 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 1.6 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.1 | 1.1 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.1 | 1.6 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 | 3.1 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 0.9 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.8 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 | 0.6 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.2 | GO:0044090 | positive regulation of vacuole organization(GO:0044090) |
| 0.0 | 0.3 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.0 | 0.4 | GO:0007314 | cytoplasm organization(GO:0007028) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) intracellular mRNA localization(GO:0008298) maternal determination of anterior/posterior axis, embryo(GO:0008358) |
| 0.0 | 0.6 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 1.1 | GO:0061009 | intrahepatic bile duct development(GO:0035622) common bile duct development(GO:0061009) |
| 0.0 | 3.3 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 1.1 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.0 | 0.5 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.0 | 0.3 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.0 | 1.2 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 3.5 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 1.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 1.3 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.0 | 0.4 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 1.0 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.5 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.8 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.5 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.5 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.3 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 1.8 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.7 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 1.8 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 3.6 | GO:0001817 | regulation of cytokine production(GO:0001817) |
| 0.0 | 0.1 | GO:0098751 | bone cell development(GO:0098751) |
| 0.0 | 0.8 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.0 | 8.3 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.8 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 2.3 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.5 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.3 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 1.0 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.3 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.4 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 3.3 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.2 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 1.0 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.3 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.6 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 2.9 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 0.6 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.7 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.2 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.0 | 0.6 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 0.8 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.0 | 3.4 | GO:0048701 | embryonic cranial skeleton morphogenesis(GO:0048701) |
| 0.0 | 3.5 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.0 | 1.5 | GO:0030218 | erythrocyte differentiation(GO:0030218) erythrocyte homeostasis(GO:0034101) |
| 0.0 | 1.4 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.0 | 0.1 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.0 | 11.6 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 1.9 | GO:0006401 | RNA catabolic process(GO:0006401) |
| 0.0 | 0.1 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.0 | 1.0 | GO:0006910 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) phagocytosis, recognition(GO:0006910) complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.9 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.2 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 0.0 | 0.4 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.9 | GO:0044744 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 1.0 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.0 | 1.1 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.0 | 1.4 | GO:0009617 | response to bacterium(GO:0009617) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 4.6 | GO:0006955 | immune response(GO:0006955) |
| 0.0 | 2.6 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.0 | 1.6 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.1 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.3 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.0 | 0.7 | GO:0006354 | DNA-templated transcription, elongation(GO:0006354) |
| 0.0 | 0.5 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.3 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.2 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 11.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.9 | 7.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.7 | 2.9 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.7 | 2.0 | GO:0005775 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.6 | 7.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.5 | 9.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.5 | 6.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.4 | 4.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.4 | 17.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.4 | 2.0 | GO:0097433 | dense body(GO:0097433) |
| 0.4 | 3.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.4 | 1.1 | GO:1990077 | primosome complex(GO:1990077) |
| 0.3 | 6.0 | GO:0042555 | MCM complex(GO:0042555) |
| 0.3 | 5.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.3 | 2.3 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.2 | 1.5 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.2 | 2.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.2 | 2.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 1.7 | GO:0071914 | prominosome(GO:0071914) |
| 0.2 | 0.5 | GO:0000941 | condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 0.2 | 1.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.2 | 9.9 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 3.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 2.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 1.9 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.6 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 0.9 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.1 | 1.8 | GO:0044545 | NSL complex(GO:0044545) |
| 0.1 | 2.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.7 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 3.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.8 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 6.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.8 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 0.6 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 5.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 2.5 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 1.4 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.1 | 4.2 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 0.5 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 3.7 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 0.8 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 3.8 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 2.9 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.6 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 2.8 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 7.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 0.6 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 2.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.7 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 0.6 | GO:0000177 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 0.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 13.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 18.4 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.3 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.7 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 2.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 4.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 4.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.4 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 1.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.7 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 9.6 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.1 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 1.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.1 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.0 | 0.2 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.3 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.5 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 2.0 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.6 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 2.3 | GO:0070161 | adherens junction(GO:0005912) anchoring junction(GO:0070161) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 1.9 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 1.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 5.7 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.9 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.2 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 1.6 | 4.7 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 1.4 | 5.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 1.3 | 6.6 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 1.3 | 6.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 1.1 | 3.4 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 1.0 | 3.0 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.8 | 2.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.7 | 3.6 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.7 | 6.4 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.7 | 2.0 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.6 | 1.8 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.6 | 1.8 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.5 | 1.5 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 0.4 | 3.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.4 | 3.0 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.4 | 2.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.4 | 4.5 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.4 | 2.0 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.4 | 4.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.4 | 4.0 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.4 | 2.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.4 | 4.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.3 | 12.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.3 | 2.4 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.3 | 4.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.3 | 1.9 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.3 | 9.9 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.3 | 2.3 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.3 | 2.6 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.3 | 4.8 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.3 | 5.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.3 | 1.8 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.3 | 1.0 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.2 | 4.0 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.2 | 1.7 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.2 | 2.6 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.2 | 1.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 0.7 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.2 | 3.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.2 | 1.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 5.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 0.8 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 2.6 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 0.8 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.2 | 1.9 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.2 | 1.3 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.2 | 4.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 0.7 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 2.1 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.2 | 8.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.2 | 0.6 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.1 | 1.0 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 1.4 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 6.3 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 2.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 1.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 1.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 2.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.7 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 1.8 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 8.2 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 0.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 1.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 2.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.6 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.1 | 0.8 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 1.0 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 1.1 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 0.5 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.1 | 0.9 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 0.7 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 1.4 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.7 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.8 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 9.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 1.0 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.8 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 0.6 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 3.0 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 0.9 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.5 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 1.3 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 1.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.6 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.5 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.1 | 4.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.9 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.1 | 0.5 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.1 | 5.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 2.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 4.4 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 4.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.7 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 2.4 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 0.8 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 1.5 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 0.5 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 0.6 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 20.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 0.5 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 3.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 3.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 1.1 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.0 | 5.8 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 5.7 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 1.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 3.1 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.9 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.1 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.2 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.6 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 5.2 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 14.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.2 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.0 | 6.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 4.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.9 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 3.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 14.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.3 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 6.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 3.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.8 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.9 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.4 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 2.3 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.7 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 4.0 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 13.1 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 1.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.3 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 1.8 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.8 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.1 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.6 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.7 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.7 | GO:0003678 | DNA helicase activity(GO:0003678) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 6.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.5 | 4.0 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.3 | 4.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.3 | 6.9 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 2.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 6.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 3.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 11.1 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.2 | 8.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 2.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 1.6 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 6.4 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 2.0 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 2.6 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 3.3 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 1.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 1.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 3.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 0.6 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 0.6 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 1.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 1.6 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 1.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 1.1 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 0.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.9 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 3.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.8 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 3.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.7 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.4 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.1 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.7 | 6.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.5 | 5.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.5 | 3.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.4 | 6.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.4 | 3.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.3 | 2.4 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.3 | 1.8 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.3 | 3.0 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.3 | 2.7 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.3 | 1.4 | REACTOME G PROTEIN BETA GAMMA SIGNALLING | Genes involved in G-protein beta:gamma signalling |
| 0.2 | 1.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 3.8 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 5.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.2 | 3.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.2 | 1.8 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.2 | 1.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 1.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 3.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 2.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 2.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.9 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.1 | 1.1 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 2.7 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 1.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 1.1 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 3.0 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 1.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 0.9 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 0.7 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 0.5 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 0.8 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.1 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.5 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.0 | 0.8 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 2.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 2.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 2.3 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.9 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |