Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
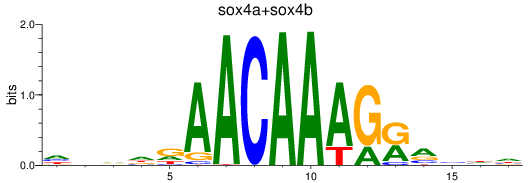
Results for sox4a+sox4b
Z-value: 0.80
Transcription factors associated with sox4a+sox4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox4a
|
ENSDARG00000004588 | SRY-box transcription factor 4a |
|
sox4b
|
ENSDARG00000098834 | SRY-box transcription factor 4b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox4b | dr11_v1_chr16_+_68069_68124 | 0.61 | 7.4e-11 | Click! |
| sox4a | dr11_v1_chr19_-_28789404_28789409 | 0.47 | 2.3e-06 | Click! |
Activity profile of sox4a+sox4b motif
Sorted Z-values of sox4a+sox4b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_30615901 | 9.17 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr16_-_9675982 | 6.51 |
ENSDART00000113724
|
mal2
|
mal, T cell differentiation protein 2 (gene/pseudogene) |
| chr14_-_8080416 | 6.31 |
ENSDART00000045109
|
zgc:92242
|
zgc:92242 |
| chr16_-_33104944 | 5.87 |
ENSDART00000151943
|
pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr13_+_22675802 | 5.47 |
ENSDART00000145538
ENSDART00000143312 |
zgc:193505
|
zgc:193505 |
| chr3_-_27646070 | 5.32 |
ENSDART00000122031
ENSDART00000151027 |
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr5_+_20823409 | 4.31 |
ENSDART00000093185
ENSDART00000142894 |
limk2
|
LIM domain kinase 2 |
| chr5_-_37117778 | 4.30 |
ENSDART00000149138
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr5_+_15203421 | 4.25 |
ENSDART00000040826
|
tbx1
|
T-box 1 |
| chr9_+_48007081 | 4.14 |
ENSDART00000060593
ENSDART00000099835 |
zgc:92380
|
zgc:92380 |
| chr17_-_5860222 | 4.10 |
ENSDART00000058894
|
si:ch73-340m8.2
|
si:ch73-340m8.2 |
| chr4_-_16354292 | 4.04 |
ENSDART00000139919
|
lum
|
lumican |
| chr8_-_39903803 | 3.93 |
ENSDART00000012391
|
cabp1a
|
calcium binding protein 1a |
| chr15_-_12360409 | 3.72 |
ENSDART00000164596
|
tmprss13a
|
transmembrane protease, serine 13a |
| chr4_+_18843015 | 3.65 |
ENSDART00000152086
ENSDART00000066977 ENSDART00000132567 |
bik
|
BCL2 interacting killer |
| chr23_-_3758637 | 3.62 |
ENSDART00000131536
ENSDART00000139408 ENSDART00000137826 |
hmga1a
|
high mobility group AT-hook 1a |
| chr17_-_2036850 | 3.41 |
ENSDART00000186048
ENSDART00000188838 ENSDART00000186423 |
spint1a
|
serine peptidase inhibitor, Kunitz type 1 a |
| chr1_+_44173245 | 3.28 |
ENSDART00000159450
ENSDART00000106048 ENSDART00000157763 |
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr19_+_7636941 | 3.18 |
ENSDART00000081611
ENSDART00000163805 ENSDART00000112404 |
cgnb
|
cingulin b |
| chr2_+_45191049 | 3.17 |
ENSDART00000165392
|
ccl20a.3
|
chemokine (C-C motif) ligand 20a, duplicate 3 |
| chr25_+_35502552 | 3.12 |
ENSDART00000189612
ENSDART00000058443 |
fibina
|
fin bud initiation factor a |
| chr7_+_20019125 | 3.08 |
ENSDART00000186391
|
bcl6b
|
B-cell CLL/lymphoma 6, member B |
| chr23_-_29824146 | 2.84 |
ENSDART00000020616
|
zgc:194189
|
zgc:194189 |
| chr22_+_18886209 | 2.79 |
ENSDART00000144402
|
fstl3
|
follistatin-like 3 (secreted glycoprotein) |
| chr8_+_46217861 | 2.78 |
ENSDART00000038790
|
angptl7
|
angiopoietin-like 7 |
| chr18_+_39327010 | 2.75 |
ENSDART00000012164
|
tmod2
|
tropomodulin 2 |
| chr23_-_3759345 | 2.73 |
ENSDART00000132205
ENSDART00000137707 ENSDART00000189382 |
hmga1a
|
high mobility group AT-hook 1a |
| chr1_+_54773877 | 2.68 |
ENSDART00000129831
|
nlrc6
|
NLR family CARD domain containing 6 |
| chr3_-_16110351 | 2.59 |
ENSDART00000064838
|
lasp1
|
LIM and SH3 protein 1 |
| chr11_+_29965822 | 2.57 |
ENSDART00000127049
|
il1fma
|
interleukin-1 family member A |
| chr12_-_22400999 | 2.55 |
ENSDART00000153194
|
si:dkey-38p12.3
|
si:dkey-38p12.3 |
| chr21_-_37027252 | 2.55 |
ENSDART00000076483
|
zgc:77151
|
zgc:77151 |
| chr17_-_8862424 | 2.53 |
ENSDART00000064633
|
nkl.4
|
NK-lysin tandem duplicate 4 |
| chr11_-_45185792 | 2.48 |
ENSDART00000171328
|
si:dkey-93h22.7
|
si:dkey-93h22.7 |
| chr13_+_28732101 | 2.48 |
ENSDART00000015773
|
ldb1a
|
LIM domain binding 1a |
| chr16_-_49505275 | 2.44 |
ENSDART00000160784
|
satb1b
|
SATB homeobox 1b |
| chr24_-_33756003 | 2.42 |
ENSDART00000079283
|
tmeff1b
|
transmembrane protein with EGF-like and two follistatin-like domains 1b |
| chr20_+_6533260 | 2.42 |
ENSDART00000135005
ENSDART00000166356 |
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr3_-_16110100 | 2.41 |
ENSDART00000051807
|
lasp1
|
LIM and SH3 protein 1 |
| chr9_+_25330905 | 2.40 |
ENSDART00000101470
|
itm2bb
|
integral membrane protein 2Bb |
| chr6_+_12968101 | 2.38 |
ENSDART00000013781
|
mcm6
|
minichromosome maintenance complex component 6 |
| chr2_+_5300405 | 2.38 |
ENSDART00000019925
|
GNB4
|
G protein subunit beta 4 |
| chr3_+_56645710 | 2.30 |
ENSDART00000193978
|
CR759836.1
|
|
| chr5_-_46505691 | 2.23 |
ENSDART00000111589
ENSDART00000122966 ENSDART00000166907 |
hapln1a
|
hyaluronan and proteoglycan link protein 1a |
| chr20_+_34770197 | 2.16 |
ENSDART00000018304
|
mcm3
|
minichromosome maintenance complex component 3 |
| chr6_-_8244474 | 2.12 |
ENSDART00000151358
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr24_+_21973929 | 2.12 |
ENSDART00000042495
|
sat1b
|
spermidine/spermine N1-acetyltransferase 1b |
| chr6_-_6976096 | 2.11 |
ENSDART00000151822
ENSDART00000039443 ENSDART00000177960 |
tuba8l4
|
tubulin, alpha 8 like 4 |
| chr16_+_29586468 | 2.09 |
ENSDART00000148926
|
mcl1b
|
MCL1, BCL2 family apoptosis regulator b |
| chr1_-_23308225 | 2.08 |
ENSDART00000137567
ENSDART00000008201 |
smim14
|
small integral membrane protein 14 |
| chr10_+_20113830 | 2.04 |
ENSDART00000139722
|
dmtn
|
dematin actin binding protein |
| chr21_+_26389391 | 2.04 |
ENSDART00000077197
|
tmsb
|
thymosin, beta |
| chr8_-_28349859 | 2.03 |
ENSDART00000062671
|
tuba8l
|
tubulin, alpha 8 like |
| chr8_+_30797363 | 2.02 |
ENSDART00000077280
|
mmp11a
|
matrix metallopeptidase 11a |
| chr18_+_30998472 | 1.99 |
ENSDART00000154993
ENSDART00000099333 |
cd151l
|
CD151 antigen, like |
| chr15_-_15469079 | 1.95 |
ENSDART00000132637
ENSDART00000004220 |
rab34a
|
RAB34, member RAS oncogene family a |
| chr7_-_28413224 | 1.95 |
ENSDART00000076502
|
rerglb
|
RERG/RAS-like b |
| chr22_-_10354381 | 1.92 |
ENSDART00000092050
|
stab1
|
stabilin 1 |
| chr8_+_4803906 | 1.89 |
ENSDART00000045533
|
tmem127
|
transmembrane protein 127 |
| chr3_-_36364903 | 1.85 |
ENSDART00000028883
|
gna13b
|
guanine nucleotide binding protein (G protein), alpha 13b |
| chr22_-_7025393 | 1.84 |
ENSDART00000003422
|
smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr19_-_15855427 | 1.82 |
ENSDART00000133059
|
cited4a
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4a |
| chr14_+_33525196 | 1.81 |
ENSDART00000085335
|
zdhhc9
|
zinc finger, DHHC-type containing 9 |
| chr1_+_44173506 | 1.80 |
ENSDART00000170512
|
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr14_-_33095917 | 1.74 |
ENSDART00000074720
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr15_+_37331585 | 1.72 |
ENSDART00000170715
|
zgc:171592
|
zgc:171592 |
| chr5_+_23045096 | 1.70 |
ENSDART00000171719
|
atrxl
|
alpha thalassemia/mental retardation syndrome X-linked, like |
| chr5_+_31480342 | 1.69 |
ENSDART00000098197
|
si:dkey-220k22.1
|
si:dkey-220k22.1 |
| chr6_+_8176486 | 1.66 |
ENSDART00000193308
|
nfil3-5
|
nuclear factor, interleukin 3 regulated, member 5 |
| chr6_+_59832786 | 1.65 |
ENSDART00000154985
ENSDART00000102148 |
ddx3b
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3b |
| chr13_-_21650404 | 1.63 |
ENSDART00000078352
|
tspan14
|
tetraspanin 14 |
| chr12_-_28848015 | 1.63 |
ENSDART00000153200
|
si:ch211-194k22.8
|
si:ch211-194k22.8 |
| chr15_+_12429206 | 1.61 |
ENSDART00000168997
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr23_+_42819221 | 1.60 |
ENSDART00000180495
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr12_+_10115964 | 1.59 |
ENSDART00000152369
|
si:dkeyp-118b1.2
|
si:dkeyp-118b1.2 |
| chr10_+_29849977 | 1.59 |
ENSDART00000180242
|
hspa8
|
heat shock protein 8 |
| chr17_+_44030692 | 1.56 |
ENSDART00000049503
|
peli2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr25_-_12902242 | 1.55 |
ENSDART00000164733
|
sept15
|
septin 15 |
| chr19_+_7424347 | 1.55 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr7_-_55454406 | 1.55 |
ENSDART00000108646
|
piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr1_-_9652547 | 1.54 |
ENSDART00000016750
ENSDART00000127735 ENSDART00000160474 ENSDART00000080323 ENSDART00000126996 ENSDART00000126877 ENSDART00000123773 |
ugt5b3
ugt5b2
ugt5b3
ugt5b1
|
UDP glucuronosyltransferase 5 family, polypeptide B3 UDP glucuronosyltransferase 5 family, polypeptide B2 UDP glucuronosyltransferase 5 family, polypeptide B3 UDP glucuronosyltransferase 5 family, polypeptide B1 |
| chr22_-_15693085 | 1.51 |
ENSDART00000141861
|
si:ch1073-396h14.1
|
si:ch1073-396h14.1 |
| chr21_+_25071805 | 1.50 |
ENSDART00000078651
|
dixdc1b
|
DIX domain containing 1b |
| chr10_-_25628555 | 1.49 |
ENSDART00000143978
|
tiam1a
|
T cell lymphoma invasion and metastasis 1a |
| chr19_-_17774875 | 1.49 |
ENSDART00000151133
ENSDART00000130695 |
top2b
|
DNA topoisomerase II beta |
| chr18_-_7481036 | 1.49 |
ENSDART00000101292
|
si:dkey-238c7.16
|
si:dkey-238c7.16 |
| chr2_+_18988407 | 1.49 |
ENSDART00000170216
|
glula
|
glutamate-ammonia ligase (glutamine synthase) a |
| chr18_+_8231138 | 1.48 |
ENSDART00000140193
|
arsa
|
arylsulfatase A |
| chr7_-_26408472 | 1.46 |
ENSDART00000111494
|
gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr12_+_48340133 | 1.46 |
ENSDART00000152899
ENSDART00000153335 ENSDART00000054788 |
ddit4
|
DNA-damage-inducible transcript 4 |
| chr21_-_32781612 | 1.45 |
ENSDART00000031028
|
cnot6a
|
CCR4-NOT transcription complex, subunit 6a |
| chr2_-_24069331 | 1.43 |
ENSDART00000156972
ENSDART00000181691 ENSDART00000157041 |
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr15_+_28368823 | 1.42 |
ENSDART00000142298
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr22_+_4443689 | 1.42 |
ENSDART00000185490
|
ticam1
|
toll-like receptor adaptor molecule 1 |
| chr12_-_20616160 | 1.39 |
ENSDART00000105362
|
snx11
|
sorting nexin 11 |
| chr1_+_54834119 | 1.39 |
ENSDART00000140020
|
si:ch211-196h16.4
|
si:ch211-196h16.4 |
| chr16_-_38001040 | 1.36 |
ENSDART00000133861
ENSDART00000138711 ENSDART00000143846 ENSDART00000146564 |
si:ch211-198c19.3
|
si:ch211-198c19.3 |
| chr15_+_28368644 | 1.34 |
ENSDART00000168453
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr9_-_8670158 | 1.31 |
ENSDART00000077296
|
col4a1
|
collagen, type IV, alpha 1 |
| chr22_-_22337382 | 1.29 |
ENSDART00000144684
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr23_-_36306337 | 1.28 |
ENSDART00000142760
ENSDART00000136929 ENSDART00000143340 |
cbx5
|
chromobox homolog 5 (HP1 alpha homolog, Drosophila) |
| chr5_+_41477954 | 1.28 |
ENSDART00000185871
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr10_-_36633882 | 1.28 |
ENSDART00000077161
ENSDART00000137688 |
rsf1b.1
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 remodeling and spacing factor 1b, tandem duplicate 1 |
| chr8_+_29636431 | 1.27 |
ENSDART00000133047
|
smarcad1a
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 a |
| chr11_-_183328 | 1.27 |
ENSDART00000168562
|
avil
|
advillin |
| chr9_+_54179306 | 1.26 |
ENSDART00000189829
|
tmsb4x
|
thymosin, beta 4 x |
| chr6_-_40768654 | 1.26 |
ENSDART00000184668
ENSDART00000146470 |
arpc4
|
actin related protein 2/3 complex, subunit 4 |
| chr1_+_24076243 | 1.25 |
ENSDART00000014608
|
mab21l2
|
mab-21-like 2 |
| chr14_+_6962271 | 1.25 |
ENSDART00000148447
ENSDART00000149114 ENSDART00000149492 ENSDART00000148394 |
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr6_-_19351495 | 1.25 |
ENSDART00000164287
|
grb2a
|
growth factor receptor-bound protein 2a |
| chr19_+_40026959 | 1.24 |
ENSDART00000123647
|
sfpq
|
splicing factor proline/glutamine-rich |
| chr5_-_32383475 | 1.23 |
ENSDART00000141294
|
fbxw2
|
F-box and WD repeat domain containing 2 |
| chr19_-_46037835 | 1.23 |
ENSDART00000163815
|
nup153
|
nucleoporin 153 |
| chr6_-_40098641 | 1.21 |
ENSDART00000017402
|
ip6k2b
|
inositol hexakisphosphate kinase 2b |
| chr6_-_9707599 | 1.21 |
ENSDART00000039280
|
fzd7b
|
frizzled class receptor 7b |
| chr19_-_30800004 | 1.21 |
ENSDART00000128560
ENSDART00000045504 ENSDART00000125893 |
trit1
|
tRNA isopentenyltransferase 1 |
| chr6_+_46406565 | 1.20 |
ENSDART00000168440
ENSDART00000131203 ENSDART00000138567 ENSDART00000132845 |
pbrm1l
|
polybromo 1, like |
| chr14_+_6963312 | 1.19 |
ENSDART00000150050
|
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr23_+_25428513 | 1.18 |
ENSDART00000144554
|
fmnl3
|
formin-like 3 |
| chr3_-_21348478 | 1.18 |
ENSDART00000114906
|
fam171a2a
|
family with sequence similarity 171, member A2a |
| chr2_-_3038904 | 1.18 |
ENSDART00000186795
|
guk1a
|
guanylate kinase 1a |
| chr17_+_20504196 | 1.17 |
ENSDART00000190539
|
neurl1ab
|
neuralized E3 ubiquitin protein ligase 1Ab |
| chr11_+_807153 | 1.13 |
ENSDART00000173289
|
vgll4b
|
vestigial-like family member 4b |
| chr22_-_22301672 | 1.13 |
ENSDART00000111711
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr22_+_28446365 | 1.12 |
ENSDART00000189359
|
abi3bpb
|
ABI family, member 3 (NESH) binding protein b |
| chr8_-_51340773 | 1.12 |
ENSDART00000060633
|
kansl3
|
KAT8 regulatory NSL complex subunit 3 |
| chr1_+_5485799 | 1.11 |
ENSDART00000022307
|
atic
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr17_+_24632440 | 1.10 |
ENSDART00000157092
|
map4k3b
|
mitogen-activated protein kinase kinase kinase kinase 3b |
| chr12_-_30777743 | 1.10 |
ENSDART00000148888
|
entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr13_+_39188737 | 1.09 |
ENSDART00000083641
|
fam135a
|
family with sequence similarity 135, member A |
| chr11_-_2250767 | 1.09 |
ENSDART00000018131
|
hnrnpa1a
|
heterogeneous nuclear ribonucleoprotein A1a |
| chr6_-_59505589 | 1.06 |
ENSDART00000170685
|
gli1
|
GLI family zinc finger 1 |
| chr13_-_33317323 | 1.04 |
ENSDART00000110295
ENSDART00000144848 ENSDART00000136701 |
tmem234
|
transmembrane protein 234 |
| chr15_-_4094888 | 1.04 |
ENSDART00000166307
|
TM4SF19
|
si:dkey-83h2.3 |
| chr2_-_7696287 | 1.04 |
ENSDART00000190769
|
CABZ01055306.1
|
|
| chr15_+_34963316 | 1.03 |
ENSDART00000153840
|
si:ch73-95l15.5
|
si:ch73-95l15.5 |
| chr5_+_41477526 | 1.03 |
ENSDART00000153567
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr20_-_33790003 | 1.03 |
ENSDART00000020183
|
fam102bb
|
family with sequence similarity 102, member B, b |
| chr10_-_2788668 | 1.02 |
ENSDART00000131749
ENSDART00000124356 ENSDART00000085031 |
ash2l
|
ash2 (absent, small, or homeotic)-like (Drosophila) |
| chr10_+_29849497 | 1.02 |
ENSDART00000099994
ENSDART00000132212 |
hspa8
|
heat shock protein 8 |
| chr25_+_34862225 | 1.02 |
ENSDART00000149782
|
CHST6
|
zgc:194879 |
| chr6_+_7322587 | 1.01 |
ENSDART00000065500
|
abcc4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
| chr3_+_52999962 | 1.01 |
ENSDART00000104683
|
pbx4
|
pre-B-cell leukemia transcription factor 4 |
| chr15_-_15468326 | 1.01 |
ENSDART00000161192
|
rab34a
|
RAB34, member RAS oncogene family a |
| chr7_-_24149670 | 1.00 |
ENSDART00000005884
|
mmp14a
|
matrix metallopeptidase 14a (membrane-inserted) |
| chr19_+_24575077 | 1.00 |
ENSDART00000167469
|
si:dkeyp-92c9.4
|
si:dkeyp-92c9.4 |
| chr3_+_3545825 | 0.99 |
ENSDART00000109060
|
CR589947.1
|
|
| chr8_-_51340931 | 0.98 |
ENSDART00000178209
|
kansl3
|
KAT8 regulatory NSL complex subunit 3 |
| chr2_+_2967255 | 0.98 |
ENSDART00000167649
ENSDART00000166449 |
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr24_+_17142881 | 0.98 |
ENSDART00000177272
ENSDART00000192259 ENSDART00000191029 |
mllt10
|
MLLT10, histone lysine methyltransferase DOT1L cofactor |
| chr14_-_31151148 | 0.98 |
ENSDART00000026569
|
gpc4
|
glypican 4 |
| chr13_+_25422009 | 0.94 |
ENSDART00000057686
|
calhm2
|
calcium homeostasis modulator 2 |
| chr22_+_12161738 | 0.94 |
ENSDART00000168857
|
ccnt2b
|
cyclin T2b |
| chr8_-_19467011 | 0.94 |
ENSDART00000162010
|
zgc:92140
|
zgc:92140 |
| chr20_+_26881600 | 0.94 |
ENSDART00000174799
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr20_-_25533739 | 0.93 |
ENSDART00000063064
|
cyp2ad6
|
cytochrome P450, family 2, subfamily AD, polypeptide 6 |
| chr5_-_38179220 | 0.93 |
ENSDART00000147701
|
si:ch211-284e13.11
|
si:ch211-284e13.11 |
| chr9_-_36924388 | 0.93 |
ENSDART00000059756
|
ralba
|
v-ral simian leukemia viral oncogene homolog Ba (ras related) |
| chr5_-_8096232 | 0.93 |
ENSDART00000158447
|
nipbla
|
nipped-B homolog a (Drosophila) |
| chr25_+_31323978 | 0.91 |
ENSDART00000067030
|
lsp1
|
lymphocyte-specific protein 1 |
| chr4_+_13953537 | 0.90 |
ENSDART00000133596
|
pphln1
|
periphilin 1 |
| chr22_+_8979955 | 0.89 |
ENSDART00000144005
|
si:ch211-213a13.1
|
si:ch211-213a13.1 |
| chr3_+_34985895 | 0.89 |
ENSDART00000121981
|
smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr5_-_22602979 | 0.89 |
ENSDART00000146287
|
nono
|
non-POU domain containing, octamer-binding |
| chr21_+_26748141 | 0.89 |
ENSDART00000169025
|
pcxa
|
pyruvate carboxylase a |
| chr23_+_9088191 | 0.88 |
ENSDART00000030811
|
cables2b
|
Cdk5 and Abl enzyme substrate 2b |
| chr24_-_35707552 | 0.88 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr10_-_1697037 | 0.87 |
ENSDART00000125188
ENSDART00000002985 |
srsf9
|
serine/arginine-rich splicing factor 9 |
| chr4_-_15003854 | 0.87 |
ENSDART00000134701
ENSDART00000002401 |
klhdc10
|
kelch domain containing 10 |
| chr3_+_60761811 | 0.85 |
ENSDART00000053482
|
tsen54
|
TSEN54 tRNA splicing endonuclease subunit |
| chr5_+_19448078 | 0.85 |
ENSDART00000088968
|
ube3b
|
ubiquitin protein ligase E3B |
| chr7_+_44713135 | 0.84 |
ENSDART00000170721
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr11_+_11152214 | 0.83 |
ENSDART00000148030
|
ly75
|
lymphocyte antigen 75 |
| chr14_-_2004291 | 0.83 |
ENSDART00000114039
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr19_-_29853402 | 0.82 |
ENSDART00000024292
ENSDART00000188508 |
txlna
|
taxilin alpha |
| chr24_-_31194847 | 0.82 |
ENSDART00000158808
|
cnn3a
|
calponin 3, acidic a |
| chr24_-_12983829 | 0.82 |
ENSDART00000133324
|
dcaf11
|
ddb1 and cul4 associated factor 11 |
| chr9_-_32142311 | 0.82 |
ENSDART00000142768
|
ankrd44
|
ankyrin repeat domain 44 |
| chr1_+_41609254 | 0.82 |
ENSDART00000132189
|
mogs
|
mannosyl-oligosaccharide glucosidase |
| chr13_+_25421531 | 0.82 |
ENSDART00000158093
|
calhm2
|
calcium homeostasis modulator 2 |
| chr22_+_14117078 | 0.81 |
ENSDART00000013575
|
bzw1a
|
basic leucine zipper and W2 domains 1a |
| chr23_-_16682186 | 0.81 |
ENSDART00000020810
|
sdcbp2
|
syndecan binding protein (syntenin) 2 |
| chr5_+_36900157 | 0.81 |
ENSDART00000183533
ENSDART00000051184 |
hnrnpl
|
heterogeneous nuclear ribonucleoprotein L |
| chr5_-_24231139 | 0.81 |
ENSDART00000143492
|
senp3a
|
SUMO1/sentrin/SMT3 specific peptidase 3a |
| chr10_-_17284055 | 0.80 |
ENSDART00000167464
|
GNAZ
|
G protein subunit alpha z |
| chr16_-_33105677 | 0.79 |
ENSDART00000145055
|
pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr24_-_6345647 | 0.78 |
ENSDART00000108994
|
zgc:174877
|
zgc:174877 |
| chr19_+_43969363 | 0.78 |
ENSDART00000051712
|
gatad1
|
GATA zinc finger domain containing 1 |
| chr6_+_7249531 | 0.78 |
ENSDART00000125912
ENSDART00000083424 ENSDART00000049695 ENSDART00000136088 |
dzip1
|
DAZ interacting zinc finger protein 1 |
| chr7_+_22801465 | 0.77 |
ENSDART00000052862
ENSDART00000173633 |
rbm4.1
|
RNA binding motif protein 4.1 |
| chr5_+_25317061 | 0.77 |
ENSDART00000170097
|
trpm6
|
transient receptor potential cation channel, subfamily M, member 6 |
| chr16_-_54919260 | 0.76 |
ENSDART00000156533
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr12_-_30777540 | 0.75 |
ENSDART00000126466
|
entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr15_-_6976851 | 0.75 |
ENSDART00000158474
ENSDART00000168943 ENSDART00000169944 |
si:ch73-311h14.2
|
si:ch73-311h14.2 |
| chr24_-_6345965 | 0.74 |
ENSDART00000178287
|
zgc:174877
|
zgc:174877 |
| chr12_+_19320657 | 0.73 |
ENSDART00000100075
ENSDART00000066389 |
tmem184ba
|
transmembrane protein 184ba |
| chr16_+_25068576 | 0.73 |
ENSDART00000125838
|
im:7147486
|
im:7147486 |
| chr12_-_20120702 | 0.73 |
ENSDART00000153387
ENSDART00000158412 ENSDART00000112768 |
ubald1a
|
UBA-like domain containing 1a |
| chr2_-_57264262 | 0.72 |
ENSDART00000183815
ENSDART00000149829 ENSDART00000088508 ENSDART00000149508 |
mbd3a
|
methyl-CpG binding domain protein 3a |
| chr22_-_33916620 | 0.72 |
ENSDART00000191276
|
CR974456.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of sox4a+sox4b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 1.3 | 6.5 | GO:0045056 | transcytosis(GO:0045056) |
| 0.6 | 2.4 | GO:1903019 | negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.5 | 2.1 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.5 | 2.0 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.5 | 1.9 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.5 | 1.4 | GO:0035666 | regulation of natural killer cell activation(GO:0032814) positive regulation of natural killer cell activation(GO:0032816) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.5 | 1.9 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.5 | 2.3 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.4 | 2.6 | GO:1902946 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.4 | 4.5 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.4 | 3.0 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.4 | 1.1 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.3 | 1.6 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.3 | 1.9 | GO:0033986 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.3 | 2.8 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.3 | 1.5 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.3 | 0.8 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.3 | 1.3 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.3 | 2.8 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.2 | 5.1 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.2 | 2.1 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.2 | 0.9 | GO:0071169 | establishment of mitotic sister chromatid cohesion(GO:0034087) establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.2 | 6.7 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.2 | 2.0 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.2 | 1.5 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.2 | 1.8 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.2 | 7.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.2 | 0.9 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.2 | 1.0 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.2 | 0.5 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.2 | 1.3 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.2 | 1.2 | GO:0070571 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.1 | 2.1 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.1 | 1.3 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.1 | 0.5 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 1.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.9 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.1 | 1.3 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 0.9 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 0.9 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.1 | 0.9 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.1 | 2.0 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 1.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.3 | GO:0051352 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.1 | 2.3 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.9 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.1 | 2.4 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 0.5 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 2.3 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 0.7 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 3.2 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 1.0 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 3.3 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 2.5 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 1.0 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 2.8 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.1 | 3.6 | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043280) |
| 0.1 | 0.8 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.7 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 1.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 1.8 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 1.0 | GO:0060729 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.5 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.1 | 1.5 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 0.3 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.5 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.1 | 1.6 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 0.8 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 1.8 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.5 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.1 | GO:0021961 | posterior commissure morphogenesis(GO:0021961) |
| 0.0 | 0.7 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 6.6 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 1.0 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 1.2 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 1.7 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 1.9 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 1.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 1.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.2 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.0 | 0.4 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 | 0.9 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.7 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 1.3 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 1.0 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.5 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 1.6 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 0.0 | 1.7 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.5 | GO:0021508 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.0 | 0.5 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 4.0 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 1.5 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 4.1 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 2.2 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.9 | GO:0070304 | positive regulation of stress-activated MAPK cascade(GO:0032874) positive regulation of stress-activated protein kinase signaling cascade(GO:0070304) |
| 0.0 | 0.4 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.7 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 2.9 | GO:0001817 | regulation of cytokine production(GO:0001817) |
| 0.0 | 1.2 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.0 | 1.1 | GO:0009311 | oligosaccharide metabolic process(GO:0009311) |
| 0.0 | 0.2 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.0 | 0.4 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.0 | 2.5 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 1.3 | GO:0071599 | otic vesicle development(GO:0071599) |
| 0.0 | 0.5 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 | 0.8 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.5 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.1 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 1.7 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.6 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 1.1 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 3.3 | GO:0006955 | immune response(GO:0006955) |
| 0.0 | 0.7 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.6 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.8 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.4 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.0 | 0.1 | GO:0014812 | muscle cell migration(GO:0014812) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.4 | 1.3 | GO:0031213 | RSF complex(GO:0031213) |
| 0.3 | 1.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.3 | 1.1 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.3 | 4.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 0.8 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.2 | 1.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 1.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.2 | 0.5 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.2 | 2.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 2.1 | GO:0044545 | NSL complex(GO:0044545) |
| 0.1 | 0.5 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 0.5 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 0.4 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.9 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 0.8 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 0.3 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 6.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 0.3 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 0.7 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 1.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 1.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 1.0 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 5.8 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.8 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 1.7 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 1.8 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 1.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 2.8 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.9 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 1.0 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.7 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 3.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 4.5 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 4.6 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 4.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 4.1 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 3.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 3.2 | GO:0043296 | bicellular tight junction(GO:0005923) apical junction complex(GO:0043296) occluding junction(GO:0070160) |
| 0.0 | 0.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 4.2 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 1.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.5 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 1.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 5.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.3 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 2.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 1.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 3.0 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.8 | GO:0031674 | I band(GO:0031674) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.5 | 1.5 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.5 | 2.4 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.4 | 6.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.4 | 1.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.4 | 2.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.3 | 1.0 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.3 | 2.1 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.3 | 0.9 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.3 | 0.8 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 1.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 1.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.2 | 1.5 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 1.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 2.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.2 | 2.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 3.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 1.5 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.2 | 0.5 | GO:0032143 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.1 | 2.8 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 2.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 1.1 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.1 | 0.5 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.7 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 1.9 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 2.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.9 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 1.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 1.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 2.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.9 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 3.2 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 1.5 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.6 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.7 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 1.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 2.4 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 0.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 4.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.8 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 1.0 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 1.1 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 1.5 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.2 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.4 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 4.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.5 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 1.8 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 4.4 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 3.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 2.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.6 | GO:0042379 | chemokine activity(GO:0008009) chemokine receptor binding(GO:0042379) |
| 0.0 | 4.5 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 1.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 1.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 1.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 9.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 5.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 10.7 | GO:0000989 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 3.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 3.7 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.7 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 1.2 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 3.7 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 2.2 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 3.9 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.1 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 5.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 4.3 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.2 | 2.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 4.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 0.8 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 1.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 1.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 0.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 1.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 3.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 3.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.3 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.1 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 2.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.2 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.3 | 4.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.3 | 4.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.3 | 1.6 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.2 | 5.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 2.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 1.4 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 2.4 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 1.5 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 0.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.1 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.1 | 1.0 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.0 | 1.6 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.9 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 1.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 1.0 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.7 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.8 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |