Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
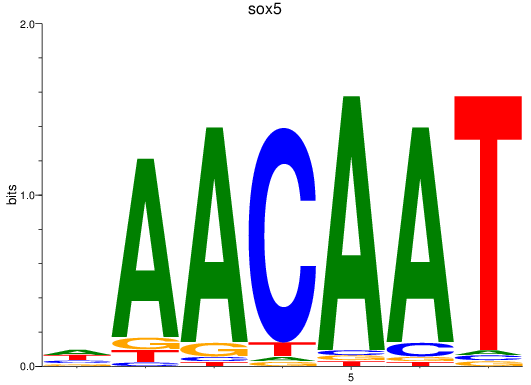
Results for sox5
Z-value: 1.42
Transcription factors associated with sox5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox5
|
ENSDARG00000011582 | SRY-box transcription factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox5 | dr11_v1_chr4_-_17055782_17055782 | 0.53 | 5.3e-08 | Click! |
Activity profile of sox5 motif
Sorted Z-values of sox5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_21832441 | 24.27 |
ENSDART00000151272
ENSDART00000151442 ENSDART00000150168 ENSDART00000148797 ENSDART00000128196 ENSDART00000149259 ENSDART00000052556 ENSDART00000149658 ENSDART00000149639 ENSDART00000148424 |
mbpa
|
myelin basic protein a |
| chr5_+_37837245 | 22.60 |
ENSDART00000171617
|
epd
|
ependymin |
| chr20_+_34915945 | 17.45 |
ENSDART00000153064
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr16_+_10318893 | 16.01 |
ENSDART00000055380
|
tubb5
|
tubulin, beta 5 |
| chr2_+_21855036 | 14.28 |
ENSDART00000140012
|
ca8
|
carbonic anhydrase VIII |
| chr13_+_7292061 | 13.34 |
ENSDART00000179504
|
CABZ01072077.1
|
Danio rerio neuroblast differentiation-associated protein AHNAK-like (LOC795051), mRNA. |
| chr12_-_3756405 | 13.19 |
ENSDART00000150839
|
fam57bb
|
family with sequence similarity 57, member Bb |
| chr10_+_22003750 | 12.84 |
ENSDART00000109420
|
kcnip1b
|
Kv channel interacting protein 1 b |
| chr3_-_35602233 | 12.59 |
ENSDART00000055269
|
gng13b
|
guanine nucleotide binding protein (G protein), gamma 13b |
| chr23_-_3681026 | 11.93 |
ENSDART00000192128
ENSDART00000040086 |
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr13_+_38520927 | 11.69 |
ENSDART00000137299
ENSDART00000111080 |
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr15_-_33933790 | 11.55 |
ENSDART00000165162
ENSDART00000182258 ENSDART00000183240 |
mag
|
myelin associated glycoprotein |
| chr11_+_39672874 | 11.48 |
ENSDART00000046663
ENSDART00000157659 |
camta1b
|
calmodulin binding transcription activator 1b |
| chr3_-_31804481 | 11.46 |
ENSDART00000028270
|
gfap
|
glial fibrillary acidic protein |
| chr15_+_28685892 | 11.02 |
ENSDART00000155815
ENSDART00000060244 |
nova2
|
neuro-oncological ventral antigen 2 |
| chr14_-_33872092 | 10.83 |
ENSDART00000111903
|
si:ch73-335m24.2
|
si:ch73-335m24.2 |
| chr20_-_47704973 | 10.68 |
ENSDART00000174808
|
tfap2b
|
transcription factor AP-2 beta |
| chr6_-_21873266 | 10.58 |
ENSDART00000151658
ENSDART00000151152 ENSDART00000151179 |
si:dkey-19e4.5
|
si:dkey-19e4.5 |
| chr5_-_50748081 | 10.57 |
ENSDART00000113985
|
mctp1a
|
multiple C2 domains, transmembrane 1a |
| chr3_-_42086577 | 10.39 |
ENSDART00000083111
ENSDART00000187312 |
ttyh3a
|
tweety family member 3a |
| chr1_-_38709551 | 10.38 |
ENSDART00000128794
|
gpm6ab
|
glycoprotein M6Ab |
| chr13_-_22771708 | 10.30 |
ENSDART00000112900
|
si:ch211-150i13.1
|
si:ch211-150i13.1 |
| chr20_-_35246150 | 10.26 |
ENSDART00000090549
|
fzd3a
|
frizzled class receptor 3a |
| chr16_-_43025885 | 10.15 |
ENSDART00000193146
ENSDART00000157302 |
si:dkey-7j14.5
|
si:dkey-7j14.5 |
| chr23_+_19590006 | 9.91 |
ENSDART00000021231
|
slmapb
|
sarcolemma associated protein b |
| chr6_+_18520859 | 9.91 |
ENSDART00000158263
|
si:dkey-10p5.10
|
si:dkey-10p5.10 |
| chr16_-_28856112 | 9.58 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr7_-_52849913 | 9.46 |
ENSDART00000174133
ENSDART00000172951 |
map1aa
|
microtubule-associated protein 1Aa |
| chr18_-_1185772 | 9.38 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr3_+_24134418 | 9.37 |
ENSDART00000156204
|
si:ch211-246i5.5
|
si:ch211-246i5.5 |
| chr7_+_47287307 | 9.05 |
ENSDART00000114669
|
dpy19l3
|
dpy-19 like C-mannosyltransferase 3 |
| chr8_+_31799410 | 8.91 |
ENSDART00000141226
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr19_+_24872159 | 8.74 |
ENSDART00000158490
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
| chr13_+_11440389 | 8.73 |
ENSDART00000186463
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr24_+_24461341 | 8.56 |
ENSDART00000147658
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr5_+_58397646 | 8.55 |
ENSDART00000180759
|
HEPACAM
|
hepatic and glial cell adhesion molecule |
| chr16_+_23811554 | 8.37 |
ENSDART00000114336
|
si:dkey-7f3.9
|
si:dkey-7f3.9 |
| chr3_+_29714775 | 8.35 |
ENSDART00000041388
|
cacng2a
|
calcium channel, voltage-dependent, gamma subunit 2a |
| chr14_-_18671334 | 8.25 |
ENSDART00000182381
|
slitrk4
|
SLIT and NTRK-like family, member 4 |
| chr24_+_24461558 | 7.92 |
ENSDART00000182424
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr5_-_46273938 | 7.82 |
ENSDART00000080033
|
si:ch211-130m23.3
|
si:ch211-130m23.3 |
| chr2_+_50626476 | 7.74 |
ENSDART00000018150
|
neurod6b
|
neuronal differentiation 6b |
| chr6_+_40587551 | 7.74 |
ENSDART00000155554
|
frs3
|
fibroblast growth factor receptor substrate 3 |
| chr15_+_18863875 | 7.67 |
ENSDART00000062603
|
cadm1b
|
cell adhesion molecule 1b |
| chr22_-_26945493 | 7.52 |
ENSDART00000077411
|
cxcl12b
|
chemokine (C-X-C motif) ligand 12b (stromal cell-derived factor 1) |
| chr25_-_29134654 | 7.44 |
ENSDART00000067066
|
parp6b
|
poly (ADP-ribose) polymerase family, member 6b |
| chr24_-_6647275 | 7.06 |
ENSDART00000161494
|
arhgap21a
|
Rho GTPase activating protein 21a |
| chr7_-_38861741 | 7.00 |
ENSDART00000173629
ENSDART00000037361 ENSDART00000173953 |
phf21aa
|
PHD finger protein 21Aa |
| chr19_+_37848830 | 6.92 |
ENSDART00000042276
ENSDART00000180872 |
nxph1
|
neurexophilin 1 |
| chr13_+_28854438 | 6.80 |
ENSDART00000193407
ENSDART00000189554 |
CU639469.1
|
|
| chr19_-_103289 | 6.80 |
ENSDART00000143118
|
adgrb1b
|
adhesion G protein-coupled receptor B1b |
| chr16_-_21047872 | 6.77 |
ENSDART00000131582
|
cbx3b
|
chromobox homolog 3b |
| chr24_+_32411753 | 6.72 |
ENSDART00000058530
|
neurod6a
|
neuronal differentiation 6a |
| chr10_-_26163989 | 6.70 |
ENSDART00000136472
|
trim3b
|
tripartite motif containing 3b |
| chr13_+_51579851 | 6.67 |
ENSDART00000163847
|
nkx6.2
|
NK6 homeobox 2 |
| chr10_+_23022263 | 6.67 |
ENSDART00000138955
|
si:dkey-175g6.2
|
si:dkey-175g6.2 |
| chr2_+_56463167 | 6.47 |
ENSDART00000123392
|
rab11bb
|
RAB11B, member RAS oncogene family, b |
| chr6_-_38418862 | 6.33 |
ENSDART00000104135
|
gabra5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr8_+_23093155 | 6.30 |
ENSDART00000063075
|
zgc:100920
|
zgc:100920 |
| chr8_-_39978767 | 6.27 |
ENSDART00000083066
|
asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr13_-_20518632 | 6.26 |
ENSDART00000165310
|
gfra1a
|
gdnf family receptor alpha 1a |
| chr9_+_32978302 | 6.25 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| chr6_+_39812475 | 6.12 |
ENSDART00000067063
|
c1ql4b
|
complement component 1, q subcomponent-like 4b |
| chr8_+_31941650 | 6.02 |
ENSDART00000138217
|
htr1aa
|
5-hydroxytryptamine (serotonin) receptor 1A a |
| chr8_-_34051548 | 5.92 |
ENSDART00000105204
|
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr17_-_39185336 | 5.86 |
ENSDART00000050534
|
crim1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
| chr23_-_30787932 | 5.86 |
ENSDART00000135771
|
myt1a
|
myelin transcription factor 1a |
| chr5_-_18446483 | 5.86 |
ENSDART00000180027
|
si:dkey-215k6.1
|
si:dkey-215k6.1 |
| chr2_+_24867534 | 5.80 |
ENSDART00000158050
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr18_-_12327426 | 5.79 |
ENSDART00000136992
ENSDART00000114024 |
fam107b
|
family with sequence similarity 107, member B |
| chr24_-_20658446 | 5.75 |
ENSDART00000127923
|
nktr
|
natural killer cell triggering receptor |
| chr17_+_33495194 | 5.75 |
ENSDART00000033691
|
pth2
|
parathyroid hormone 2 |
| chr19_-_15335787 | 5.71 |
ENSDART00000187131
|
hivep3a
|
human immunodeficiency virus type I enhancer binding protein 3a |
| chr8_-_9118958 | 5.64 |
ENSDART00000037922
|
slc6a8
|
solute carrier family 6 (neurotransmitter transporter), member 8 |
| chr19_+_15571290 | 5.61 |
ENSDART00000131134
|
foxo6b
|
forkhead box O6 b |
| chr17_+_19626479 | 5.60 |
ENSDART00000044993
ENSDART00000131863 |
rgs7a
|
regulator of G protein signaling 7a |
| chr2_-_21170517 | 5.58 |
ENSDART00000135417
|
bmi1b
|
bmi1 polycomb ring finger oncogene 1b |
| chr22_-_16443199 | 5.56 |
ENSDART00000006290
ENSDART00000193335 |
plekhb2
|
pleckstrin homology domain containing, family B (evectins) member 2 |
| chr23_+_30730121 | 5.54 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr11_+_25010491 | 5.54 |
ENSDART00000167285
|
zgc:92107
|
zgc:92107 |
| chr22_-_20342260 | 5.52 |
ENSDART00000161610
ENSDART00000165667 |
tcf3b
|
transcription factor 3b |
| chr11_+_6819050 | 5.48 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr10_+_21807497 | 5.46 |
ENSDART00000164634
|
pcdh1g32
|
protocadherin 1 gamma 32 |
| chr7_+_23515966 | 5.45 |
ENSDART00000186893
ENSDART00000186189 |
zgc:109889
|
zgc:109889 |
| chr1_-_30689004 | 5.38 |
ENSDART00000018827
|
dachc
|
dachshund c |
| chr17_-_33289304 | 5.38 |
ENSDART00000135118
ENSDART00000040346 |
efr3ba
|
EFR3 homolog Ba (S. cerevisiae) |
| chr2_+_31957554 | 5.37 |
ENSDART00000012413
|
ankhb
|
ANKH inorganic pyrophosphate transport regulator b |
| chr17_+_28005763 | 5.35 |
ENSDART00000155838
|
luzp1
|
leucine zipper protein 1 |
| chr23_-_36003282 | 5.31 |
ENSDART00000103150
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr6_+_42819337 | 5.27 |
ENSDART00000046498
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr8_+_25267903 | 5.23 |
ENSDART00000093090
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr15_-_20949692 | 5.23 |
ENSDART00000185548
|
tbcela
|
tubulin folding cofactor E-like a |
| chr18_-_21267103 | 5.22 |
ENSDART00000136285
|
pnp6
|
purine nucleoside phosphorylase 6 |
| chr13_+_12045758 | 5.15 |
ENSDART00000079398
ENSDART00000165467 ENSDART00000165880 |
gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr1_+_9708801 | 5.12 |
ENSDART00000189621
|
elfn1b
|
extracellular leucine-rich repeat and fibronectin type III domain containing 1b |
| chr17_-_14966384 | 5.09 |
ENSDART00000105064
|
txndc16
|
thioredoxin domain containing 16 |
| chr16_-_22006996 | 5.09 |
ENSDART00000116114
|
si:dkey-71b5.7
|
si:dkey-71b5.7 |
| chr9_+_42607138 | 5.09 |
ENSDART00000138133
ENSDART00000002027 |
gulp1a
|
GULP, engulfment adaptor PTB domain containing 1a |
| chr20_-_14206698 | 5.06 |
ENSDART00000082441
|
dlgap2b
|
discs, large (Drosophila) homolog-associated protein 2b |
| chr2_-_36819624 | 5.04 |
ENSDART00000140844
|
slitrk3b
|
SLIT and NTRK-like family, member 3b |
| chr11_+_16152316 | 4.92 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr5_+_41793001 | 4.90 |
ENSDART00000136439
ENSDART00000190477 ENSDART00000192289 |
bcl7a
|
BCL tumor suppressor 7A |
| chr6_+_11250316 | 4.89 |
ENSDART00000137122
|
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr16_+_5678071 | 4.80 |
ENSDART00000011166
ENSDART00000134198 ENSDART00000131575 |
zgc:158689
|
zgc:158689 |
| chr9_+_31534601 | 4.72 |
ENSDART00000133094
|
si:ch211-168k14.2
|
si:ch211-168k14.2 |
| chr10_-_28761454 | 4.63 |
ENSDART00000129400
|
alcama
|
activated leukocyte cell adhesion molecule a |
| chr5_-_28606916 | 4.62 |
ENSDART00000026107
ENSDART00000137717 |
tnc
|
tenascin C |
| chr19_+_43969363 | 4.60 |
ENSDART00000051712
|
gatad1
|
GATA zinc finger domain containing 1 |
| chr14_+_31529958 | 4.58 |
ENSDART00000053026
|
fam122b
|
family with sequence similarity 122B |
| chr15_-_33834577 | 4.57 |
ENSDART00000163354
|
mmp13b
|
matrix metallopeptidase 13b |
| chr3_+_54047342 | 4.56 |
ENSDART00000178486
|
olfm2a
|
olfactomedin 2a |
| chr3_+_40407352 | 4.52 |
ENSDART00000155112
|
tnrc18
|
trinucleotide repeat containing 18 |
| chr13_+_12045475 | 4.50 |
ENSDART00000163053
ENSDART00000160812 ENSDART00000158244 |
gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr20_+_27331008 | 4.49 |
ENSDART00000141486
|
ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr8_+_13106760 | 4.44 |
ENSDART00000029308
|
itgb4
|
integrin, beta 4 |
| chr20_+_54383838 | 4.39 |
ENSDART00000157737
|
lrfn5b
|
leucine rich repeat and fibronectin type III domain containing 5b |
| chr6_+_12326267 | 4.36 |
ENSDART00000155101
|
si:dkey-276j7.3
|
si:dkey-276j7.3 |
| chr19_+_7549854 | 4.31 |
ENSDART00000138866
ENSDART00000151758 |
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr1_-_22726233 | 4.19 |
ENSDART00000140920
|
prom1b
|
prominin 1 b |
| chr10_+_33744098 | 4.15 |
ENSDART00000147775
|
rxfp2a
|
relaxin/insulin-like family peptide receptor 2a |
| chr13_-_20519001 | 4.13 |
ENSDART00000168955
|
gfra1a
|
gdnf family receptor alpha 1a |
| chr17_-_21418340 | 4.12 |
ENSDART00000007021
|
atp6v1ba
|
ATPase, H+ transporting, lysosomal, V1 subunit B, member a |
| chr17_-_24564674 | 4.03 |
ENSDART00000105435
ENSDART00000135086 |
abch1
|
ATP-binding cassette, sub-family H, member 1 |
| chr5_-_32158198 | 4.01 |
ENSDART00000187215
ENSDART00000137503 |
m17
|
IL-6 subfamily cytokine M17 |
| chr13_+_29773153 | 3.99 |
ENSDART00000144443
ENSDART00000133796 ENSDART00000141310 ENSDART00000139782 |
pax2a
|
paired box 2a |
| chr18_+_38900092 | 3.96 |
ENSDART00000148541
ENSDART00000133618 ENSDART00000088037 |
myo5aa
|
myosin VAa |
| chr6_+_54358529 | 3.92 |
ENSDART00000153704
|
anks1ab
|
ankyrin repeat and sterile alpha motif domain containing 1Ab |
| chr16_+_27614989 | 3.90 |
ENSDART00000005625
|
glipr2l
|
GLI pathogenesis-related 2, like |
| chr16_+_19033554 | 3.90 |
ENSDART00000182430
|
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr4_-_19016396 | 3.90 |
ENSDART00000166160
|
si:dkey-31f5.11
|
si:dkey-31f5.11 |
| chr11_+_21587950 | 3.90 |
ENSDART00000162251
|
FAM72B
|
zgc:101564 |
| chr5_-_38342992 | 3.88 |
ENSDART00000140337
|
mink1
|
misshapen-like kinase 1 |
| chr7_-_18601206 | 3.86 |
ENSDART00000111636
|
DTX4
|
si:ch211-119e14.2 |
| chr7_+_25003851 | 3.85 |
ENSDART00000144526
|
naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae) |
| chr23_+_20523617 | 3.85 |
ENSDART00000176404
|
adnpb
|
activity-dependent neuroprotector homeobox b |
| chr5_+_20147830 | 3.84 |
ENSDART00000098727
|
svopa
|
SV2 related protein a |
| chr6_+_42818963 | 3.83 |
ENSDART00000184833
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr7_+_30970045 | 3.83 |
ENSDART00000155974
|
tjp1a
|
tight junction protein 1a |
| chr16_-_21047483 | 3.81 |
ENSDART00000136235
|
cbx3b
|
chromobox homolog 3b |
| chr24_+_26039464 | 3.77 |
ENSDART00000131017
|
tnk2a
|
tyrosine kinase, non-receptor, 2a |
| chr21_-_25573064 | 3.75 |
ENSDART00000134310
|
CR388166.1
|
|
| chr14_+_49683767 | 3.72 |
ENSDART00000192515
|
tspan17
|
tetraspanin 17 |
| chr9_-_33749556 | 3.72 |
ENSDART00000149383
|
ndp
|
Norrie disease (pseudoglioma) |
| chr19_+_1964005 | 3.72 |
ENSDART00000172049
|
sh3bp5a
|
SH3-domain binding protein 5a (BTK-associated) |
| chr10_+_15255012 | 3.72 |
ENSDART00000023766
|
vldlr
|
very low density lipoprotein receptor |
| chr2_+_16696052 | 3.70 |
ENSDART00000022356
ENSDART00000164329 |
ppp1r7
|
protein phosphatase 1, regulatory (inhibitor) subunit 7 |
| chr1_+_45958904 | 3.69 |
ENSDART00000108528
|
arhgef7b
|
Rho guanine nucleotide exchange factor (GEF) 7b |
| chr18_+_19131773 | 3.65 |
ENSDART00000060766
|
rab11a
|
RAB11a, member RAS oncogene family |
| chr10_+_15255198 | 3.63 |
ENSDART00000139047
ENSDART00000172107 ENSDART00000183413 ENSDART00000185314 |
vldlr
|
very low density lipoprotein receptor |
| chr23_-_33944597 | 3.61 |
ENSDART00000133223
|
si:dkey-190g6.2
|
si:dkey-190g6.2 |
| chr4_-_8902406 | 3.61 |
ENSDART00000192962
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr21_-_27201495 | 3.55 |
ENSDART00000145013
|
zgc:77262
|
zgc:77262 |
| chr22_+_18530395 | 3.54 |
ENSDART00000105415
ENSDART00000183958 |
si:ch211-212d10.1
|
si:ch211-212d10.1 |
| chr5_-_40910749 | 3.50 |
ENSDART00000083467
ENSDART00000133183 |
parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr13_+_3667230 | 3.50 |
ENSDART00000131553
ENSDART00000189841 ENSDART00000183554 ENSDART00000018737 |
qkib
|
QKI, KH domain containing, RNA binding b |
| chr13_+_3938526 | 3.50 |
ENSDART00000012759
|
yipf3
|
Yip1 domain family, member 3 |
| chr11_+_37612214 | 3.46 |
ENSDART00000172899
ENSDART00000077496 |
hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr11_+_16153207 | 3.45 |
ENSDART00000192356
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr9_-_27649406 | 3.40 |
ENSDART00000181270
ENSDART00000187112 |
stxbp5l
|
syntaxin binding protein 5-like |
| chr3_+_52999962 | 3.40 |
ENSDART00000104683
|
pbx4
|
pre-B-cell leukemia transcription factor 4 |
| chr20_-_31238313 | 3.40 |
ENSDART00000028471
|
hpcal1
|
hippocalcin-like 1 |
| chr6_-_28943056 | 3.39 |
ENSDART00000065138
|
tbc1d23
|
TBC1 domain family, member 23 |
| chr19_-_45960191 | 3.33 |
ENSDART00000052434
ENSDART00000172732 |
eif3hb
|
eukaryotic translation initiation factor 3, subunit H, b |
| chr15_+_31471808 | 3.32 |
ENSDART00000110078
|
or102-3
|
odorant receptor, family C, subfamily 102, member 3 |
| chr11_+_37612672 | 3.30 |
ENSDART00000157993
|
hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr24_+_17069420 | 3.28 |
ENSDART00000014787
|
pip4k2aa
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha a |
| chr21_+_37357578 | 3.27 |
ENSDART00000143621
|
nsd1b
|
nuclear receptor binding SET domain protein 1b |
| chr22_-_29906764 | 3.25 |
ENSDART00000019786
|
smc3
|
structural maintenance of chromosomes 3 |
| chr2_+_38556195 | 3.24 |
ENSDART00000138769
|
cdh24b
|
cadherin 24, type 2b |
| chr7_-_31938938 | 3.22 |
ENSDART00000132353
|
bdnf
|
brain-derived neurotrophic factor |
| chr25_-_29134000 | 3.20 |
ENSDART00000172027
ENSDART00000190447 |
parp6b
|
poly (ADP-ribose) polymerase family, member 6b |
| chr14_-_2217285 | 3.19 |
ENSDART00000157949
ENSDART00000166150 ENSDART00000054891 ENSDART00000183268 |
pcdh2ab2
pcdh2ab2
|
protocadherin 2 alpha b2 protocadherin 2 alpha b2 |
| chr19_+_20793388 | 3.14 |
ENSDART00000142463
|
txnl4a
|
thioredoxin-like 4A |
| chr23_-_30727596 | 3.12 |
ENSDART00000060193
|
thap3
|
THAP domain containing, apoptosis associated protein 3 |
| chr4_-_16412084 | 3.10 |
ENSDART00000188460
|
dcn
|
decorin |
| chr21_-_2707768 | 3.10 |
ENSDART00000165384
|
CABZ01101739.1
|
|
| chr14_-_30808174 | 3.10 |
ENSDART00000173262
|
prss23
|
protease, serine, 23 |
| chr9_+_38163876 | 3.09 |
ENSDART00000137955
|
clasp1a
|
cytoplasmic linker associated protein 1a |
| chr6_-_12900154 | 3.09 |
ENSDART00000080408
ENSDART00000150887 |
ical1
|
islet cell autoantigen 1-like |
| chr25_+_388258 | 3.08 |
ENSDART00000166834
|
rfx7b
|
regulatory factor X7b |
| chr19_-_38872650 | 3.07 |
ENSDART00000146641
|
adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr15_-_21669618 | 3.06 |
ENSDART00000156995
|
sorl1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr21_+_10782013 | 3.04 |
ENSDART00000146248
|
znf532
|
zinc finger protein 532 |
| chr5_-_67911111 | 3.03 |
ENSDART00000051833
|
gsx1
|
GS homeobox 1 |
| chr15_+_29728377 | 3.03 |
ENSDART00000099958
|
zgc:153372
|
zgc:153372 |
| chr5_+_17624463 | 3.02 |
ENSDART00000183869
ENSDART00000081064 |
fbrsl1
|
fibrosin-like 1 |
| chr23_-_36446307 | 2.98 |
ENSDART00000136623
|
zgc:174906
|
zgc:174906 |
| chr15_+_5028608 | 2.92 |
ENSDART00000092809
|
abcg1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr20_+_38837238 | 2.91 |
ENSDART00000061334
|
ift172
|
intraflagellar transport 172 |
| chr2_-_36818132 | 2.89 |
ENSDART00000110447
|
slitrk3b
|
SLIT and NTRK-like family, member 3b |
| chr13_+_13770980 | 2.87 |
ENSDART00000113089
|
slc4a11
|
solute carrier family 4, sodium borate transporter, member 11 |
| chr17_+_23968214 | 2.86 |
ENSDART00000183053
|
xpo1b
|
exportin 1 (CRM1 homolog, yeast) b |
| chr17_-_35278763 | 2.83 |
ENSDART00000063437
|
adam17a
|
ADAM metallopeptidase domain 17a |
| chr6_+_9163007 | 2.81 |
ENSDART00000183054
ENSDART00000157552 |
zgc:112023
|
zgc:112023 |
| chr6_+_32382743 | 2.81 |
ENSDART00000190009
|
dock7
|
dedicator of cytokinesis 7 |
| chr9_+_35860975 | 2.79 |
ENSDART00000134447
|
rcan1a
|
regulator of calcineurin 1a |
| chr2_-_48966431 | 2.73 |
ENSDART00000147948
|
kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr15_-_14467394 | 2.71 |
ENSDART00000191944
|
numbl
|
numb homolog (Drosophila)-like |
| chr20_+_26943072 | 2.69 |
ENSDART00000153215
|
cdca4
|
cell division cycle associated 4 |
| chr14_-_24761132 | 2.68 |
ENSDART00000146299
|
slit3
|
slit homolog 3 (Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.3 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 3.0 | 9.1 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 2.3 | 11.7 | GO:0016322 | neuron remodeling(GO:0016322) |
| 1.9 | 7.5 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) |
| 1.8 | 8.9 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 1.7 | 6.8 | GO:0090386 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 1.5 | 4.6 | GO:0045887 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 1.2 | 3.7 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 1.1 | 12.6 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 1.1 | 3.4 | GO:1990403 | embryonic brain development(GO:1990403) |
| 1.0 | 13.5 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.8 | 6.6 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.8 | 2.4 | GO:1902895 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.7 | 2.9 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.7 | 2.1 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.7 | 7.7 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.6 | 5.6 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.6 | 2.5 | GO:0071548 | cellular response to cortisol stimulus(GO:0071387) response to dexamethasone(GO:0071548) |
| 0.6 | 4.3 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.6 | 17.9 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.6 | 3.4 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.6 | 4.5 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.6 | 11.7 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.5 | 11.9 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.5 | 3.7 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.5 | 1.6 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.5 | 1.6 | GO:0046831 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.5 | 2.6 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.5 | 8.3 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.5 | 4.9 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.5 | 2.4 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.5 | 5.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.4 | 3.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.4 | 1.3 | GO:1900182 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.4 | 6.2 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.4 | 2.0 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.4 | 6.8 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.4 | 2.3 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.4 | 5.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.4 | 14.1 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.4 | 1.5 | GO:0015840 | urea transport(GO:0015840) |
| 0.4 | 9.5 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.4 | 1.8 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.3 | 3.8 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.3 | 12.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.3 | 2.0 | GO:1902915 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.3 | 3.9 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.3 | 4.1 | GO:0045762 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.3 | 2.9 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.3 | 3.1 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.3 | 3.7 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.3 | 19.9 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.3 | 8.6 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.3 | 3.9 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.3 | 5.2 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.3 | 3.7 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.3 | 8.4 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.3 | 12.8 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.3 | 6.3 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.3 | 6.0 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.3 | 12.1 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.3 | 3.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 9.7 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.2 | 32.1 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.2 | 4.0 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.2 | 5.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.2 | 6.0 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.2 | 1.7 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.2 | 2.3 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.2 | 1.4 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.2 | 13.7 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.2 | 1.3 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.2 | 3.6 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.2 | 1.7 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.2 | 2.8 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.2 | 5.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 2.7 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.2 | 4.8 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.2 | 6.1 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.2 | 3.5 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.2 | 17.0 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.2 | 2.1 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.2 | 5.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 1.4 | GO:2000047 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) regulation of cell-cell adhesion mediated by cadherin(GO:2000047) |
| 0.2 | 15.6 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.2 | 5.2 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.2 | 0.8 | GO:0001961 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.1 | 4.0 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.1 | 4.6 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 4.4 | GO:0001763 | morphogenesis of a branching structure(GO:0001763) |
| 0.1 | 3.1 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.1 | 4.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 2.9 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 0.7 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.1 | 4.4 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.1 | 3.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 2.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 1.0 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 6.8 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.1 | 0.7 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.1 | 1.0 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 7.2 | GO:0045055 | regulated exocytosis(GO:0045055) |
| 0.1 | 1.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 2.8 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 7.3 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.1 | 1.0 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 5.6 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 0.8 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 2.1 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 3.5 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.1 | 1.9 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.9 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 3.4 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.1 | 0.8 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 2.7 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 14.0 | GO:0000398 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.1 | 4.0 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.1 | 1.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 0.9 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 2.6 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 1.4 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.5 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 0.3 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 1.7 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 5.7 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 5.6 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.1 | 2.7 | GO:1990573 | potassium ion import(GO:0010107) potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 1.8 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 3.4 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 1.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 8.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 5.0 | GO:0072659 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 3.7 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.9 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.0 | 1.8 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 6.8 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.8 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.4 | GO:0099565 | excitatory postsynaptic potential(GO:0060079) chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.0 | 1.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.6 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.0 | 1.9 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.9 | GO:0048634 | regulation of muscle organ development(GO:0048634) |
| 0.0 | 1.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 1.3 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 2.4 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.9 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.4 | GO:0071453 | cellular response to decreased oxygen levels(GO:0036294) cellular response to oxygen levels(GO:0071453) cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.9 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.3 | GO:0039020 | pronephric nephron tubule development(GO:0039020) |
| 0.0 | 3.4 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 17.8 | GO:0030182 | neuron differentiation(GO:0030182) |
| 0.0 | 0.6 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 2.2 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.1 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 1.4 | GO:0051169 | nucleocytoplasmic transport(GO:0006913) nuclear transport(GO:0051169) |
| 0.0 | 0.2 | GO:0098781 | ncRNA transcription(GO:0098781) |
| 0.0 | 0.1 | GO:0097324 | adult behavior(GO:0030534) melanocyte migration(GO:0097324) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 17.5 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 1.9 | 11.7 | GO:0043083 | synaptic cleft(GO:0043083) |
| 1.6 | 28.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 1.3 | 4.0 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 1.0 | 3.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.9 | 8.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.9 | 10.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.8 | 5.5 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.7 | 3.4 | GO:0071203 | WASH complex(GO:0071203) |
| 0.6 | 3.7 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.6 | 9.4 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.5 | 7.3 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.5 | 1.6 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.5 | 4.2 | GO:0071914 | prominosome(GO:0071914) |
| 0.5 | 2.0 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.4 | 2.6 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.4 | 6.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.3 | 2.4 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.3 | 3.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.3 | 3.2 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.3 | 12.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.3 | 16.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 4.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.2 | 1.7 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 9.1 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.2 | 8.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 2.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.2 | 4.9 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.2 | 1.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 0.5 | GO:0031213 | RSF complex(GO:0031213) |
| 0.2 | 3.8 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.2 | 2.9 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 36.0 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.1 | 2.5 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 2.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 5.4 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.1 | 7.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 11.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.9 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 8.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 4.4 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 9.5 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 6.5 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 13.6 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 1.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 6.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 4.5 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.1 | 1.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 3.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 6.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 4.6 | GO:0031674 | I band(GO:0031674) |
| 0.0 | 0.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 1.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 2.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 10.4 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 1.3 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 10.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 1.3 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 7.6 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 2.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 4.2 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 1.8 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 3.6 | GO:0036477 | dendrite(GO:0030425) somatodendritic compartment(GO:0036477) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 3.3 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 7.0 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.9 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 20.4 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 2.6 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 2.3 | GO:0045202 | synapse(GO:0045202) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.3 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 1.8 | 8.9 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 1.6 | 24.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 1.5 | 10.6 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 1.3 | 7.7 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 1.2 | 10.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 1.1 | 5.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 1.1 | 5.4 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 1.0 | 22.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 1.0 | 3.9 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 0.8 | 3.3 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.7 | 5.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.7 | 2.1 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.7 | 6.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.6 | 2.6 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.6 | 11.6 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.6 | 9.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.6 | 5.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.5 | 14.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.5 | 5.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.5 | 1.4 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.5 | 2.8 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.5 | 10.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.5 | 7.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.5 | 6.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.4 | 17.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.4 | 5.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.4 | 2.0 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.4 | 3.6 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.4 | 5.8 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.4 | 5.7 | GO:0030546 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.4 | 7.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.3 | 3.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.3 | 2.0 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.3 | 2.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.3 | 9.1 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.3 | 2.9 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.3 | 16.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.3 | 14.1 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.3 | 8.6 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.3 | 1.8 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.3 | 6.0 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.2 | 3.2 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.2 | 1.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.2 | 3.3 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 4.2 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.2 | 5.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 2.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 3.3 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.2 | 5.9 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 3.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.2 | 9.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 8.4 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.2 | 2.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 1.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 6.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 6.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.2 | 1.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 1.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 3.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 3.7 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.1 | 1.9 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 2.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 9.2 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.1 | 2.9 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 6.8 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 0.5 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.9 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.8 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 1.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 2.9 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 1.7 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 3.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 4.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 7.8 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 26.7 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 4.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.2 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.1 | 1.7 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 2.1 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 1.5 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 2.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 2.2 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 8.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 2.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 21.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 1.3 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.1 | 5.0 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 0.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 4.6 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.1 | 1.0 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 4.5 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 25.5 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 1.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 16.5 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 10.6 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 1.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 2.2 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 10.9 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 29.8 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 4.0 | GO:0005126 | cytokine receptor binding(GO:0005126) |
| 0.0 | 0.1 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.0 | 1.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 8.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.7 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 5.4 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 4.1 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.0 | 1.2 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.3 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 1.2 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 1.3 | GO:0060090 | binding, bridging(GO:0060090) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 9.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.4 | 3.7 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.3 | 7.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.3 | 4.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.3 | 1.4 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.2 | 3.9 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 7.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 14.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.2 | 1.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 8.2 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.2 | 2.7 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 1.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 2.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 4.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 2.9 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 2.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 2.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 12.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.5 | 1.4 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.4 | 0.4 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.4 | 6.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.4 | 9.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.4 | 2.7 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.3 | 7.7 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.3 | 2.6 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.3 | 1.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.2 | 2.9 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 3.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 5.0 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 1.0 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 5.7 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.2 | 1.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.2 | 3.9 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 3.7 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 1.6 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 1.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 3.9 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 6.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 4.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 3.3 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 2.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 2.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 1.5 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 1.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 0.7 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 1.4 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 1.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 2.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 3.6 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |