Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
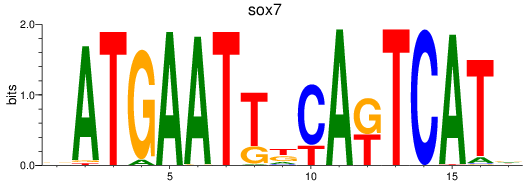
Results for sox7
Z-value: 0.73
Transcription factors associated with sox7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox7
|
ENSDARG00000030125 | SRY-box transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox7 | dr11_v1_chr20_+_19066858_19066858 | 0.63 | 1.2e-11 | Click! |
Activity profile of sox7 motif
Sorted Z-values of sox7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_29204960 | 19.46 |
ENSDART00000131386
|
pvalb7
|
parvalbumin 7 |
| chr22_-_29191152 | 19.24 |
ENSDART00000132702
|
pvalb7
|
parvalbumin 7 |
| chr22_-_29205327 | 12.68 |
ENSDART00000183161
ENSDART00000189515 |
pvalb7
|
parvalbumin 7 |
| chr15_-_23647078 | 9.75 |
ENSDART00000059366
|
ckmb
|
creatine kinase, muscle b |
| chr25_-_22639133 | 6.77 |
ENSDART00000073583
|
islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr2_-_24996441 | 5.99 |
ENSDART00000144795
|
slc35g2a
|
solute carrier family 35, member G2a |
| chr17_+_15534815 | 5.74 |
ENSDART00000159426
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr20_-_25709247 | 5.46 |
ENSDART00000146711
|
si:dkeyp-117h8.2
|
si:dkeyp-117h8.2 |
| chr16_+_46111849 | 5.43 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr20_+_50956369 | 5.23 |
ENSDART00000170854
|
gphnb
|
gephyrin b |
| chr16_+_10777116 | 4.48 |
ENSDART00000190902
|
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr14_+_6546516 | 4.01 |
ENSDART00000097214
|
adam19b
|
ADAM metallopeptidase domain 19b |
| chr9_-_23033818 | 3.87 |
ENSDART00000022392
|
rnd3b
|
Rho family GTPase 3b |
| chr22_-_12862415 | 3.86 |
ENSDART00000145156
ENSDART00000137280 |
glsa
|
glutaminase a |
| chr19_-_11031145 | 3.70 |
ENSDART00000151375
ENSDART00000027598 ENSDART00000137865 ENSDART00000188025 |
tpm3
|
tropomyosin 3 |
| chr8_+_23174137 | 3.19 |
ENSDART00000189470
|
dnajc5aa
|
DnaJ (Hsp40) homolog, subfamily C, member 5aa |
| chr9_+_38369872 | 3.18 |
ENSDART00000193914
|
plcd4b
|
phospholipase C, delta 4b |
| chr5_+_34982318 | 2.98 |
ENSDART00000160504
|
ankra2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr21_-_18932761 | 2.77 |
ENSDART00000140129
|
med15
|
mediator complex subunit 15 |
| chr14_-_6365223 | 2.74 |
ENSDART00000167446
|
slc44a1b
|
solute carrier family 44 (choline transporter), member 1b |
| chr11_-_35970966 | 2.65 |
ENSDART00000188805
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr25_-_7999756 | 2.60 |
ENSDART00000159908
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr4_+_47178124 | 2.24 |
ENSDART00000170816
|
si:dkey-26m3.2
|
si:dkey-26m3.2 |
| chr22_-_26558166 | 2.24 |
ENSDART00000111125
|
glis2a
|
GLIS family zinc finger 2a |
| chr5_-_69923241 | 2.22 |
ENSDART00000187389
|
fktn
|
fukutin |
| chr1_-_10669436 | 2.14 |
ENSDART00000152421
|
si:dkey-31e10.5
|
si:dkey-31e10.5 |
| chr18_-_39200557 | 2.07 |
ENSDART00000132367
ENSDART00000183672 |
si:ch211-235f12.2
mapk6
|
si:ch211-235f12.2 mitogen-activated protein kinase 6 |
| chr24_-_20652473 | 1.99 |
ENSDART00000179984
|
nktr
|
natural killer cell triggering receptor |
| chr1_-_470812 | 1.97 |
ENSDART00000192527
|
zgc:92518
|
zgc:92518 |
| chr19_-_42462491 | 1.96 |
ENSDART00000131715
|
psmb4
|
proteasome subunit beta 4 |
| chr8_+_2787042 | 1.88 |
ENSDART00000155275
|
sh3glb2a
|
SH3-domain GRB2-like endophilin B2a |
| chr21_-_41147818 | 1.87 |
ENSDART00000167339
ENSDART00000192730 |
msx2b
|
muscle segment homeobox 2b |
| chr11_+_44356707 | 1.78 |
ENSDART00000165219
|
srsf7b
|
serine/arginine-rich splicing factor 7b |
| chr2_-_27385934 | 1.74 |
ENSDART00000139886
|
toe1
|
target of EGR1, exonuclease |
| chr16_-_31933740 | 1.73 |
ENSDART00000125411
|
si:ch1073-90m23.1
|
si:ch1073-90m23.1 |
| chr12_-_4540564 | 1.72 |
ENSDART00000106566
|
CABZ01030107.1
|
|
| chr14_-_30490465 | 1.71 |
ENSDART00000173107
|
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr15_-_35252522 | 1.68 |
ENSDART00000144153
ENSDART00000059195 |
mff
|
mitochondrial fission factor |
| chr17_-_22010668 | 1.63 |
ENSDART00000031998
|
slc22a7b.1
|
solute carrier family 22 (organic anion transporter), member 7b, tandem duplicate 1 |
| chr8_+_16726386 | 1.59 |
ENSDART00000144621
|
smim15
|
small integral membrane protein 15 |
| chr8_+_35664152 | 1.56 |
ENSDART00000144520
|
si:dkeyp-14d3.1
|
si:dkeyp-14d3.1 |
| chr19_+_10860485 | 1.55 |
ENSDART00000169962
|
si:ch73-347e22.4
|
si:ch73-347e22.4 |
| chr13_+_24829996 | 1.40 |
ENSDART00000147194
|
si:ch211-223p8.8
|
si:ch211-223p8.8 |
| chr8_-_46457233 | 1.36 |
ENSDART00000113214
|
sult1st7
|
sulfotransferase family 1, cytosolic sulfotransferase 7 |
| chr22_-_16658996 | 1.35 |
ENSDART00000138429
|
si:dkey-38n4.2
|
si:dkey-38n4.2 |
| chr16_+_27345383 | 1.34 |
ENSDART00000078250
ENSDART00000162857 |
nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr13_+_10799380 | 1.26 |
ENSDART00000126427
|
lrpprc
|
leucine-rich pentatricopeptide repeat containing |
| chr20_-_4766645 | 1.25 |
ENSDART00000147071
ENSDART00000152398 |
ak7a
|
adenylate kinase 7a |
| chr21_+_19330774 | 1.20 |
ENSDART00000109412
|
helq
|
helicase, POLQ like |
| chr21_+_31811202 | 1.14 |
ENSDART00000189846
|
si:ch211-12m10.1
|
si:ch211-12m10.1 |
| chr8_-_40205712 | 1.06 |
ENSDART00000158927
|
anapc5
|
anaphase promoting complex subunit 5 |
| chr8_-_22514918 | 1.01 |
ENSDART00000021514
ENSDART00000189272 |
apex2
|
APEX nuclease (apurinic/apyrimidinic endonuclease) 2 |
| chr11_+_10541258 | 1.01 |
ENSDART00000132365
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr18_+_302796 | 1.00 |
ENSDART00000157434
ENSDART00000160279 |
lsm14ab
|
LSM14A mRNA processing body assembly factor b |
| chr10_-_23379146 | 0.89 |
ENSDART00000056369
|
cadm2a
|
cell adhesion molecule 2a |
| chr15_-_21692630 | 0.88 |
ENSDART00000039865
|
sdhdb
|
succinate dehydrogenase complex, subunit D, integral membrane protein b |
| chr5_-_67911111 | 0.88 |
ENSDART00000051833
|
gsx1
|
GS homeobox 1 |
| chr7_-_4107423 | 0.87 |
ENSDART00000172971
|
zgc:55733
|
zgc:55733 |
| chr22_+_6740039 | 0.80 |
ENSDART00000144122
|
CT583625.2
|
|
| chr11_+_14104417 | 0.80 |
ENSDART00000059752
ENSDART00000186575 |
med16
|
mediator complex subunit 16 |
| chr16_+_40024883 | 0.75 |
ENSDART00000110100
|
hint3
|
histidine triad nucleotide binding protein 3 |
| chr11_-_3613558 | 0.75 |
ENSDART00000163578
|
dnajc16l
|
DnaJ (Hsp40) homolog, subfamily C, member 16 like |
| chr3_+_52737565 | 0.73 |
ENSDART00000108639
|
gmip
|
GEM interacting protein |
| chr24_+_35787629 | 0.70 |
ENSDART00000136721
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr7_+_41421998 | 0.69 |
ENSDART00000083967
|
chpf2
|
chondroitin polymerizing factor 2 |
| chr2_+_27386617 | 0.64 |
ENSDART00000134976
|
si:ch73-382f3.1
|
si:ch73-382f3.1 |
| chr22_+_6843363 | 0.51 |
ENSDART00000126044
|
FO904898.3
|
|
| chr7_-_35083184 | 0.46 |
ENSDART00000100253
ENSDART00000135250 ENSDART00000173511 |
agrp
|
agouti related neuropeptide |
| chr8_+_36270571 | 0.42 |
ENSDART00000187505
|
CU929676.1
|
|
| chr14_+_1124409 | 0.41 |
ENSDART00000190043
ENSDART00000106708 ENSDART00000160246 |
sec24b
|
SEC24 homolog B, COPII coat complex component |
| chr5_-_12824607 | 0.40 |
ENSDART00000190310
|
p2rx2
|
purinergic receptor P2X, ligand-gated ion channel, 2 |
| chr4_-_77216726 | 0.30 |
ENSDART00000099943
|
psmb10
|
proteasome subunit beta 10 |
| chr16_-_31985965 | 0.30 |
ENSDART00000161944
|
si:dkey-40m6.14
|
si:dkey-40m6.14 |
| chr10_-_4961923 | 0.26 |
ENSDART00000050177
ENSDART00000146066 |
snx30
|
sorting nexin family member 30 |
| chr2_-_51511494 | 0.25 |
ENSDART00000161392
ENSDART00000160877 |
si:ch211-9d9.8
|
si:ch211-9d9.8 |
| chr8_-_21058354 | 0.25 |
ENSDART00000130481
ENSDART00000113749 |
zgc:112962
|
zgc:112962 |
| chr21_-_19330333 | 0.21 |
ENSDART00000101462
|
mrps18c
|
mitochondrial ribosomal protein S18C |
| chr2_-_39090258 | 0.19 |
ENSDART00000134270
ENSDART00000145268 |
NMNAT3
|
si:ch73-170l17.1 |
| chr9_+_11587629 | 0.16 |
ENSDART00000102508
ENSDART00000146585 |
si:dkey-69o16.5
|
si:dkey-69o16.5 |
| chr2_-_51611344 | 0.14 |
ENSDART00000158137
|
si:ch211-9d9.8
|
si:ch211-9d9.8 |
| chr7_-_61288882 | 0.12 |
ENSDART00000098616
ENSDART00000137762 |
rwdd
|
RWD domain containing 4 |
| chr9_+_32860345 | 0.11 |
ENSDART00000121751
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr12_+_26621906 | 0.09 |
ENSDART00000158440
ENSDART00000046959 |
arhgap12b
|
Rho GTPase activating protein 12b |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0018315 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.7 | 9.7 | GO:0006599 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.5 | 3.9 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.4 | 2.2 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.4 | 4.3 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.2 | 4.5 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.2 | 0.7 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.2 | 0.9 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 1.3 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 4.0 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 1.0 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 1.7 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 3.7 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.1 | 6.8 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.1 | 3.2 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 1.1 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 2.6 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.1 | 2.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 1.7 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 2.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.6 | GO:0032196 | transposition(GO:0032196) |
| 0.1 | 0.5 | GO:2000253 | adult feeding behavior(GO:0008343) positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 1.4 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.7 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 3.9 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.0 | 0.4 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 4.7 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 1.0 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 1.3 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.0 | 1.7 | GO:0090630 | activation of GTPase activity(GO:0090630) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 1.7 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 2.6 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 1.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 5.5 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 2.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 5.4 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 1.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 3.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 3.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.1 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 5.2 | GO:0099572 | postsynaptic specialization(GO:0099572) |
| 0.0 | 0.4 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.8 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.7 | 9.7 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.4 | 4.5 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.4 | 3.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.3 | 1.0 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.3 | 1.3 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.2 | 2.6 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 1.0 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.2 | 1.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 2.0 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 1.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 2.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.5 | GO:0031781 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.1 | 0.9 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 3.2 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 0.7 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 4.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 2.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 2.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 1.7 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 51.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 5.7 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.4 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 1.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 3.9 | GO:0019901 | protein kinase binding(GO:0019901) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 1.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.3 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 1.1 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 3.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |