Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
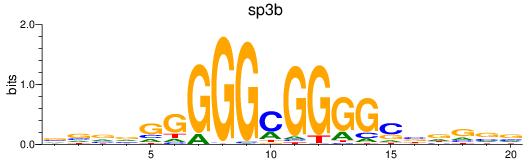
Results for sp3b
Z-value: 1.02
Transcription factors associated with sp3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sp3b
|
ENSDARG00000007812 | Sp3b transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sp3b | dr11_v1_chr6_-_10740365_10740365 | -0.55 | 7.6e-09 | Click! |
Activity profile of sp3b motif
Sorted Z-values of sp3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_1211242 | 15.88 |
ENSDART00000171287
ENSDART00000165769 |
poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr24_-_36593876 | 12.41 |
ENSDART00000160901
|
CABZ01055365.1
|
|
| chr6_+_102506 | 11.89 |
ENSDART00000172678
|
ldlrb
|
low density lipoprotein receptor b |
| chr17_-_125091 | 11.68 |
ENSDART00000158825
|
actc1b
|
actin, alpha, cardiac muscle 1b |
| chr1_-_59232267 | 11.33 |
ENSDART00000169658
ENSDART00000163257 |
akap8l
|
A kinase (PRKA) anchor protein 8-like |
| chr3_-_40276057 | 10.40 |
ENSDART00000132225
ENSDART00000074737 |
shmt1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr4_-_77557279 | 10.20 |
ENSDART00000180113
|
AL935186.10
|
|
| chr12_+_46634736 | 10.06 |
ENSDART00000008009
|
trim16
|
tripartite motif containing 16 |
| chr22_+_1006573 | 9.60 |
ENSDART00000123458
|
pparda
|
peroxisome proliferator-activated receptor delta a |
| chr24_+_38671054 | 8.99 |
ENSDART00000154214
|
si:ch73-70c5.1
|
si:ch73-70c5.1 |
| chr22_+_396840 | 8.76 |
ENSDART00000163198
|
capzb
|
capping protein (actin filament) muscle Z-line, beta |
| chr10_+_10788811 | 8.72 |
ENSDART00000101077
ENSDART00000139143 |
ptgdsa
|
prostaglandin D2 synthase a |
| chr21_+_28445052 | 8.14 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr21_-_11646878 | 8.12 |
ENSDART00000162426
ENSDART00000135937 ENSDART00000131448 ENSDART00000148097 ENSDART00000133443 |
cast
|
calpastatin |
| chr6_-_60104628 | 8.03 |
ENSDART00000057463
ENSDART00000169188 |
pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr13_+_41022502 | 7.90 |
ENSDART00000026808
|
dkk1a
|
dickkopf WNT signaling pathway inhibitor 1a |
| chr3_-_50139860 | 7.47 |
ENSDART00000101563
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr8_+_6576940 | 7.38 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr6_+_3334710 | 7.33 |
ENSDART00000132848
|
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr23_-_45407631 | 7.33 |
ENSDART00000148484
ENSDART00000150186 |
zgc:101853
|
zgc:101853 |
| chr3_-_32541033 | 7.19 |
ENSDART00000151476
ENSDART00000055324 |
rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr6_+_41503854 | 7.13 |
ENSDART00000136538
ENSDART00000140108 ENSDART00000084861 |
cish
|
cytokine inducible SH2-containing protein |
| chr24_+_35564668 | 7.01 |
ENSDART00000122734
|
cebpd
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr7_-_12968689 | 6.93 |
ENSDART00000173115
ENSDART00000013690 |
rplp2l
|
ribosomal protein, large P2, like |
| chr1_-_59216197 | 6.87 |
ENSDART00000062426
|
lpar2b
|
lysophosphatidic acid receptor 2b |
| chr18_-_6803424 | 6.85 |
ENSDART00000142647
|
si:dkey-266m15.5
|
si:dkey-266m15.5 |
| chr20_-_21672970 | 6.82 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr20_-_147574 | 6.79 |
ENSDART00000104762
ENSDART00000131635 |
slc16a10
|
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr19_+_48117995 | 6.76 |
ENSDART00000170865
|
nme2b.1
|
NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 1 |
| chr19_+_48111285 | 6.74 |
ENSDART00000169420
|
nme2b.2
|
NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 2 |
| chr3_+_32526799 | 6.61 |
ENSDART00000185755
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr3_-_18030938 | 6.61 |
ENSDART00000013540
|
si:ch73-141c7.1
|
si:ch73-141c7.1 |
| chr4_-_77551860 | 6.55 |
ENSDART00000188176
|
AL935186.6
|
|
| chr9_+_13999620 | 6.29 |
ENSDART00000143229
|
cd28l
|
cd28-like molecule |
| chr5_+_66132394 | 6.27 |
ENSDART00000073892
|
zgc:114041
|
zgc:114041 |
| chr22_-_193234 | 6.24 |
ENSDART00000131067
|
fbxo42
|
F-box protein 42 |
| chr4_+_55758103 | 6.17 |
ENSDART00000185964
|
CT583728.23
|
|
| chr8_-_31107537 | 6.00 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| chr5_-_1047222 | 5.96 |
ENSDART00000181112
|
mbd2
|
methyl-CpG binding domain protein 2 |
| chr10_-_22150419 | 5.89 |
ENSDART00000006173
|
cldn7b
|
claudin 7b |
| chr4_-_77563411 | 5.74 |
ENSDART00000186841
|
AL935186.8
|
|
| chr25_+_37443194 | 5.54 |
ENSDART00000163178
ENSDART00000190262 |
slc10a3
|
solute carrier family 10, member 3 |
| chr12_-_4540564 | 5.53 |
ENSDART00000106566
|
CABZ01030107.1
|
|
| chr21_+_10577527 | 5.39 |
ENSDART00000165070
ENSDART00000127963 |
ccbe1
|
collagen and calcium binding EGF domains 1 |
| chr20_-_54564018 | 5.33 |
ENSDART00000099832
|
zgc:153012
|
zgc:153012 |
| chr4_-_68569527 | 5.29 |
ENSDART00000192091
|
BX548011.5
|
|
| chr8_-_53490376 | 5.26 |
ENSDART00000158789
|
chdh
|
choline dehydrogenase |
| chr16_+_46294337 | 5.17 |
ENSDART00000040769
|
nr2f5
|
nuclear receptor subfamily 2, group F, member 5 |
| chr7_-_16598212 | 5.16 |
ENSDART00000128488
|
e2f8
|
E2F transcription factor 8 |
| chr13_-_31647323 | 5.15 |
ENSDART00000135381
|
six4a
|
SIX homeobox 4a |
| chr5_-_1047504 | 5.11 |
ENSDART00000159346
|
mbd2
|
methyl-CpG binding domain protein 2 |
| chr23_-_13875252 | 5.07 |
ENSDART00000104834
ENSDART00000193807 |
g6pd
|
glucose-6-phosphate dehydrogenase |
| chr15_+_19838458 | 5.05 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr12_-_45349849 | 5.02 |
ENSDART00000183036
|
CABZ01068367.1
|
Danio rerio uncharacterized LOC100332446 (LOC100332446), mRNA. |
| chr22_-_17729778 | 4.99 |
ENSDART00000192132
|
si:ch73-63e15.2
|
si:ch73-63e15.2 |
| chr13_+_22480496 | 4.95 |
ENSDART00000136863
ENSDART00000131870 ENSDART00000078720 ENSDART00000078740 ENSDART00000139218 |
ldb3a
|
LIM domain binding 3a |
| chr11_+_329687 | 4.87 |
ENSDART00000172882
|
cyp27b1
|
cytochrome P450, family 27, subfamily B, polypeptide 1 |
| chr7_+_49664174 | 4.86 |
ENSDART00000137059
ENSDART00000131210 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr3_+_32526263 | 4.84 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr6_+_3334392 | 4.83 |
ENSDART00000133707
ENSDART00000130879 |
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr3_+_29469283 | 4.81 |
ENSDART00000103592
|
fam83fa
|
family with sequence similarity 83, member Fa |
| chr3_+_5575313 | 4.75 |
ENSDART00000134693
ENSDART00000101807 |
si:ch211-106h11.3
|
si:ch211-106h11.3 |
| chr7_+_48870496 | 4.73 |
ENSDART00000009642
|
igf2a
|
insulin-like growth factor 2a |
| chr18_-_50799510 | 4.68 |
ENSDART00000174373
|
taldo1
|
transaldolase 1 |
| chr19_+_48117388 | 4.65 |
ENSDART00000160549
|
nme2b.1
|
NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 1 |
| chr18_-_26715655 | 4.59 |
ENSDART00000181497
|
malt3
|
MALT paracaspase 3 |
| chr1_+_15062 | 4.58 |
ENSDART00000169005
|
cep97
|
centrosomal protein 97 |
| chr22_-_7050 | 4.56 |
ENSDART00000127829
|
atad3
|
ATPase family, AAA domain containing 3 |
| chr14_-_2602445 | 4.56 |
ENSDART00000166910
|
etf1a
|
eukaryotic translation termination factor 1a |
| chr22_+_883678 | 4.55 |
ENSDART00000140588
|
stk38b
|
serine/threonine kinase 38b |
| chr17_-_2584423 | 4.53 |
ENSDART00000013506
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr7_-_66877058 | 4.46 |
ENSDART00000155954
|
adma
|
adrenomedullin a |
| chr12_-_48671612 | 4.46 |
ENSDART00000007202
|
zgc:92749
|
zgc:92749 |
| chr20_+_15982482 | 4.41 |
ENSDART00000020999
|
angptl1a
|
angiopoietin-like 1a |
| chr19_+_636886 | 4.41 |
ENSDART00000149192
|
tert
|
telomerase reverse transcriptase |
| chr9_+_21401189 | 4.37 |
ENSDART00000062669
|
cx30.3
|
connexin 30.3 |
| chr23_-_44848961 | 4.37 |
ENSDART00000136839
|
wu:fb72h05
|
wu:fb72h05 |
| chr4_+_1530287 | 4.32 |
ENSDART00000067446
|
slc38a4
|
solute carrier family 38, member 4 |
| chr25_+_150570 | 4.26 |
ENSDART00000170892
|
adam10b
|
ADAM metallopeptidase domain 10b |
| chr22_-_188102 | 4.26 |
ENSDART00000125391
ENSDART00000170463 |
CABZ01052268.1
|
|
| chr6_-_8580857 | 4.25 |
ENSDART00000138858
ENSDART00000041142 |
myh11a
|
myosin, heavy chain 11a, smooth muscle |
| chr3_+_26813058 | 4.24 |
ENSDART00000055537
|
socs1a
|
suppressor of cytokine signaling 1a |
| chr18_-_26715156 | 4.24 |
ENSDART00000142043
|
malt3
|
MALT paracaspase 3 |
| chr20_-_40370736 | 4.23 |
ENSDART00000041229
|
fabp7b
|
fatty acid binding protein 7, brain, b |
| chr4_+_77981553 | 4.21 |
ENSDART00000174108
ENSDART00000122459 |
terfa
|
telomeric repeat binding factor a |
| chr19_+_156757 | 4.17 |
ENSDART00000167717
|
carmil1
|
capping protein regulator and myosin 1 linker 1 |
| chr25_+_4750972 | 4.10 |
ENSDART00000168903
|
si:zfos-2372e4.1
|
si:zfos-2372e4.1 |
| chr5_-_2721686 | 4.09 |
ENSDART00000169404
|
hspa5
|
heat shock protein 5 |
| chr21_-_45891262 | 4.05 |
ENSDART00000169816
|
galnt10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) |
| chr21_-_45086170 | 4.02 |
ENSDART00000188963
|
rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr8_+_10305400 | 4.01 |
ENSDART00000172400
|
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr5_+_72152813 | 3.97 |
ENSDART00000149910
|
abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr15_+_11644866 | 3.97 |
ENSDART00000188716
|
slc1a5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr1_+_51475094 | 3.96 |
ENSDART00000146352
|
meis1a
|
Meis homeobox 1 a |
| chr21_+_1647990 | 3.95 |
ENSDART00000148540
|
fech
|
ferrochelatase |
| chr19_+_4892281 | 3.95 |
ENSDART00000150969
|
cdk12
|
cyclin-dependent kinase 12 |
| chr5_+_38200807 | 3.93 |
ENSDART00000100769
|
hsd20b2
|
hydroxysteroid (20-beta) dehydrogenase 2 |
| chr1_-_58562129 | 3.88 |
ENSDART00000159070
|
slc27a1b
|
solute carrier family 27 (fatty acid transporter), member 1b |
| chr6_+_36821621 | 3.86 |
ENSDART00000104157
|
tmem45a
|
transmembrane protein 45a |
| chr16_+_29650698 | 3.85 |
ENSDART00000137153
|
tmod4
|
tropomodulin 4 (muscle) |
| chr25_-_10724947 | 3.83 |
ENSDART00000184490
|
lrp5
|
low density lipoprotein receptor-related protein 5 |
| chr21_-_5056812 | 3.82 |
ENSDART00000139713
ENSDART00000140859 |
zgc:77838
|
zgc:77838 |
| chr23_+_44349252 | 3.80 |
ENSDART00000097644
|
ephb4b
|
eph receptor B4b |
| chr25_+_16356083 | 3.79 |
ENSDART00000125925
ENSDART00000125444 |
tead1a
|
TEA domain family member 1a |
| chr6_-_10788065 | 3.77 |
ENSDART00000190968
|
wipf1b
|
WAS/WASL interacting protein family, member 1b |
| chr8_-_53488832 | 3.73 |
ENSDART00000191801
|
chdh
|
choline dehydrogenase |
| chr9_-_8314028 | 3.73 |
ENSDART00000102739
|
si:ch211-145c1.1
|
si:ch211-145c1.1 |
| chr13_+_37022601 | 3.72 |
ENSDART00000131800
ENSDART00000041300 |
esr2b
|
estrogen receptor 2b |
| chr3_-_61162750 | 3.71 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr22_-_607812 | 3.67 |
ENSDART00000145983
|
cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr24_-_4450238 | 3.65 |
ENSDART00000066835
|
fzd8a
|
frizzled class receptor 8a |
| chr25_+_3306620 | 3.64 |
ENSDART00000182085
ENSDART00000034704 |
slc25a3b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3b |
| chr25_+_1732838 | 3.64 |
ENSDART00000159555
ENSDART00000168161 |
FBLN1
|
fibulin 1 |
| chr12_-_49168398 | 3.63 |
ENSDART00000186608
|
FO704624.1
|
|
| chr22_+_26443235 | 3.62 |
ENSDART00000044085
|
zgc:92480
|
zgc:92480 |
| chr6_-_59942335 | 3.58 |
ENSDART00000168416
|
fbxl3b
|
F-box and leucine-rich repeat protein 3b |
| chr4_-_2380173 | 3.58 |
ENSDART00000177727
|
nap1l1
|
nucleosome assembly protein 1-like 1 |
| chr15_-_33527631 | 3.55 |
ENSDART00000187646
ENSDART00000161151 |
stard13b
|
StAR-related lipid transfer (START) domain containing 13b |
| chr23_+_44374041 | 3.51 |
ENSDART00000136056
|
ephb4b
|
eph receptor B4b |
| chr24_+_17005647 | 3.49 |
ENSDART00000149149
|
zfx
|
zinc finger protein, X-linked |
| chr2_-_58075414 | 3.48 |
ENSDART00000161920
|
nectin4
|
nectin cell adhesion molecule 4 |
| chr7_+_13988075 | 3.48 |
ENSDART00000186812
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr8_-_51293265 | 3.42 |
ENSDART00000127875
ENSDART00000181145 |
bmp1a
|
bone morphogenetic protein 1a |
| chr5_-_22969424 | 3.42 |
ENSDART00000143869
ENSDART00000172549 |
si:dkey-237j10.2
|
si:dkey-237j10.2 |
| chr20_-_7080427 | 3.40 |
ENSDART00000140166
ENSDART00000023677 |
si:ch211-121a2.2
|
si:ch211-121a2.2 |
| chr12_+_48841419 | 3.35 |
ENSDART00000125331
|
dlg5b.1
|
discs, large homolog 5b (Drosophila), tandem duplicate 1 |
| chr13_-_51922290 | 3.34 |
ENSDART00000168648
|
srfb
|
serum response factor b |
| chr18_-_127558 | 3.33 |
ENSDART00000149556
|
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr25_+_4635355 | 3.33 |
ENSDART00000021120
|
pgghg
|
protein-glucosylgalactosylhydroxylysine glucosidase |
| chr16_-_53919115 | 3.29 |
ENSDART00000179533
|
fzd1
|
frizzled class receptor 1 |
| chr16_-_29387215 | 3.29 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr20_+_26556174 | 3.28 |
ENSDART00000138492
|
irf4b
|
interferon regulatory factor 4b |
| chr6_-_442163 | 3.26 |
ENSDART00000163564
ENSDART00000189134 ENSDART00000169789 |
grap2b
|
GRB2-related adaptor protein 2b |
| chr25_+_3306858 | 3.24 |
ENSDART00000137077
|
slc25a3b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3b |
| chr12_+_48841182 | 3.23 |
ENSDART00000109315
ENSDART00000185609 ENSDART00000187217 |
dlg5b.1
|
discs, large homolog 5b (Drosophila), tandem duplicate 1 |
| chr11_-_40504170 | 3.23 |
ENSDART00000165394
|
si:dkeyp-61b2.1
|
si:dkeyp-61b2.1 |
| chr10_+_187760 | 3.22 |
ENSDART00000161007
ENSDART00000160651 |
ets2
|
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
| chr23_+_19655301 | 3.21 |
ENSDART00000104441
ENSDART00000135269 |
abhd6b
|
abhydrolase domain containing 6b |
| chr9_+_1654284 | 3.21 |
ENSDART00000062854
|
nfe2l2a
|
nuclear factor, erythroid 2-like 2a |
| chr1_-_57501299 | 3.20 |
ENSDART00000080600
|
zgc:171470
|
zgc:171470 |
| chr8_+_51958968 | 3.17 |
ENSDART00000139509
ENSDART00000186544 |
gna14a
|
guanine nucleotide binding protein (G protein), alpha 14a |
| chr4_-_4612116 | 3.16 |
ENSDART00000130601
|
CABZ01020840.1
|
Danio rerio apoptosis facilitator Bcl-2-like protein 14 (LOC101885512), mRNA. |
| chr11_-_497680 | 3.15 |
ENSDART00000154888
|
cnbpb
|
CCHC-type zinc finger, nucleic acid binding protein b |
| chr23_-_31645760 | 3.12 |
ENSDART00000035031
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr2_-_47620806 | 3.08 |
ENSDART00000038228
|
ap1s3b
|
adaptor-related protein complex 1, sigma 3 subunit, b |
| chr3_+_12322170 | 3.04 |
ENSDART00000161227
|
glis2b
|
GLIS family zinc finger 2b |
| chr15_-_20468302 | 3.04 |
ENSDART00000018514
|
dlc
|
deltaC |
| chr20_+_39250673 | 3.03 |
ENSDART00000153003
|
reps1
|
RALBP1 associated Eps domain containing 1 |
| chr21_-_32436679 | 3.03 |
ENSDART00000076974
|
gfpt2
|
glutamine-fructose-6-phosphate transaminase 2 |
| chr19_+_935565 | 3.03 |
ENSDART00000113368
|
RNF5
|
ring finger protein 5 |
| chr5_-_69621227 | 3.02 |
ENSDART00000178543
|
aldh2.2
|
aldehyde dehydrogenase 2 family (mitochondrial), tandem duplicate 2 |
| chr3_+_22273123 | 3.01 |
ENSDART00000044157
|
scn4ab
|
sodium channel, voltage-gated, type IV, alpha, b |
| chr19_+_48251273 | 3.01 |
ENSDART00000157424
|
CU693379.1
|
|
| chr13_-_17860307 | 2.97 |
ENSDART00000135920
ENSDART00000054579 |
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr19_-_82504 | 2.96 |
ENSDART00000027864
ENSDART00000160560 |
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr1_+_59538755 | 2.95 |
ENSDART00000166354
|
sp6
|
Sp6 transcription factor |
| chr19_-_6193448 | 2.91 |
ENSDART00000151405
|
erf
|
Ets2 repressor factor |
| chr22_-_10752471 | 2.90 |
ENSDART00000081191
|
sass6
|
SAS-6 centriolar assembly protein |
| chr22_-_5918670 | 2.86 |
ENSDART00000141373
|
si:rp71-36a1.1
|
si:rp71-36a1.1 |
| chr1_-_58561963 | 2.86 |
ENSDART00000165040
|
slc27a1b
|
solute carrier family 27 (fatty acid transporter), member 1b |
| chr6_+_48206535 | 2.84 |
ENSDART00000075172
|
cttnbp2nla
|
CTTNBP2 N-terminal like a |
| chr10_+_44924684 | 2.84 |
ENSDART00000181360
ENSDART00000170418 ENSDART00000170327 |
sec14l7
|
SEC14-like lipid binding 7 |
| chr2_+_48073972 | 2.83 |
ENSDART00000186442
|
klf6b
|
Kruppel-like factor 6b |
| chr25_-_34284144 | 2.82 |
ENSDART00000188109
|
gcnt3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr20_-_22378401 | 2.82 |
ENSDART00000011135
ENSDART00000110967 |
kita
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog a |
| chr23_+_45456490 | 2.81 |
ENSDART00000036631
|
cyr61
|
cysteine-rich, angiogenic inducer, 61 |
| chr3_+_2669813 | 2.78 |
ENSDART00000014205
|
CR388047.1
|
|
| chr1_-_54107321 | 2.78 |
ENSDART00000148382
ENSDART00000150357 |
rfx1b
|
regulatory factor X, 1b (influences HLA class II expression) |
| chr12_-_3773869 | 2.77 |
ENSDART00000092983
|
si:ch211-166g5.4
|
si:ch211-166g5.4 |
| chr20_-_182841 | 2.76 |
ENSDART00000064546
|
si:ch211-241j12.3
|
si:ch211-241j12.3 |
| chr17_-_4395373 | 2.75 |
ENSDART00000015923
|
klhl10a
|
kelch-like family member 10a |
| chr6_-_43677125 | 2.74 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr17_-_20558961 | 2.72 |
ENSDART00000155993
|
sh3pxd2ab
|
SH3 and PX domains 2Ab |
| chr1_-_54100988 | 2.70 |
ENSDART00000192662
|
rfx1b
|
regulatory factor X, 1b (influences HLA class II expression) |
| chr2_+_55365727 | 2.68 |
ENSDART00000162943
|
FP245456.1
|
|
| chr24_+_42131564 | 2.68 |
ENSDART00000153854
|
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr25_+_25464630 | 2.67 |
ENSDART00000150537
|
rassf7a
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7a |
| chr13_+_45582391 | 2.67 |
ENSDART00000058093
|
ldlrap1b
|
low density lipoprotein receptor adaptor protein 1b |
| chr6_+_584632 | 2.65 |
ENSDART00000151150
|
zgc:92360
|
zgc:92360 |
| chr14_+_2487672 | 2.64 |
ENSDART00000170629
ENSDART00000123063 |
fgf18a
|
fibroblast growth factor 18a |
| chr24_+_37449893 | 2.62 |
ENSDART00000164405
|
nlrc3
|
NLR family, CARD domain containing 3 |
| chr15_-_17010358 | 2.61 |
ENSDART00000156768
|
hip1
|
huntingtin interacting protein 1 |
| chr3_+_49043917 | 2.60 |
ENSDART00000158212
|
zgc:92161
|
zgc:92161 |
| chr16_+_26732086 | 2.55 |
ENSDART00000138496
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr10_+_22034477 | 2.55 |
ENSDART00000133304
ENSDART00000134189 ENSDART00000021240 ENSDART00000100526 |
npm1a
|
nucleophosmin 1a |
| chr4_+_5537101 | 2.48 |
ENSDART00000008692
|
si:dkey-14d8.7
|
si:dkey-14d8.7 |
| chr18_-_41161828 | 2.47 |
ENSDART00000114993
|
CABZ01005876.1
|
|
| chr3_+_36972586 | 2.45 |
ENSDART00000102784
|
si:ch211-18i17.2
|
si:ch211-18i17.2 |
| chr4_-_23759192 | 2.43 |
ENSDART00000014685
ENSDART00000131690 |
cpm
|
carboxypeptidase M |
| chr17_-_114121 | 2.43 |
ENSDART00000172408
ENSDART00000157784 |
arhgap11a
|
Rho GTPase activating protein 11A |
| chr1_+_144284 | 2.42 |
ENSDART00000064061
|
prozb
|
protein Z, vitamin K-dependent plasma glycoprotein b |
| chr20_-_54381034 | 2.41 |
ENSDART00000136779
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr2_+_258698 | 2.40 |
ENSDART00000181330
ENSDART00000181645 |
phlpp1
|
PH domain and leucine rich repeat protein phosphatase 1 |
| chr25_+_235992 | 2.39 |
ENSDART00000123990
ENSDART00000165507 ENSDART00000153616 |
pus7
|
pseudouridylate synthase 7 (putative) |
| chr22_-_4769140 | 2.38 |
ENSDART00000165235
|
calr3a
|
calreticulin 3a |
| chr14_+_30910114 | 2.38 |
ENSDART00000187166
ENSDART00000078187 |
foxo4
|
forkhead box O4 |
| chr22_-_367569 | 2.37 |
ENSDART00000041895
|
ssu72
|
SSU72 homolog, RNA polymerase II CTD phosphatase |
| chr12_+_10443785 | 2.35 |
ENSDART00000029133
|
snu13b
|
SNU13 homolog, small nuclear ribonucleoprotein b (U4/U6.U5) |
| chr7_-_55454406 | 2.35 |
ENSDART00000108646
|
piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr8_+_29963024 | 2.33 |
ENSDART00000149372
|
ptch1
|
patched 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of sp3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.4 | GO:0019264 | L-serine catabolic process(GO:0006565) glycine biosynthetic process from serine(GO:0019264) |
| 3.0 | 9.0 | GO:0019695 | choline metabolic process(GO:0019695) |
| 2.7 | 8.1 | GO:0052575 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 2.1 | 4.2 | GO:1902745 | positive regulation of lamellipodium organization(GO:1902745) |
| 1.8 | 7.1 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 1.8 | 8.8 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 1.6 | 11.4 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 1.5 | 4.4 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 1.3 | 6.7 | GO:0071071 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 1.3 | 3.8 | GO:0035676 | anterior lateral line neuromast hair cell development(GO:0035676) |
| 1.2 | 3.6 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 1.2 | 8.1 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 1.1 | 8.0 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 1.1 | 3.4 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 1.1 | 18.2 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 1.1 | 9.6 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 1.1 | 4.2 | GO:0032208 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 1.0 | 5.9 | GO:0060876 | semicircular canal formation(GO:0060876) |
| 0.9 | 2.8 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.9 | 4.6 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.8 | 2.4 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.8 | 5.4 | GO:0060855 | venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.8 | 2.3 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.7 | 3.0 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.7 | 5.0 | GO:0021885 | forebrain cell migration(GO:0021885) |
| 0.7 | 2.8 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.7 | 3.4 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.7 | 3.3 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.6 | 1.9 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.6 | 3.1 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.6 | 1.7 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.5 | 2.1 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.5 | 1.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.5 | 3.0 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.5 | 1.4 | GO:1902102 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.5 | 4.2 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.4 | 1.3 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.4 | 3.8 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.4 | 7.9 | GO:1990399 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.4 | 2.9 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.4 | 5.0 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.4 | 1.2 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.4 | 2.9 | GO:2000463 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.4 | 1.6 | GO:0032530 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.4 | 5.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.4 | 3.7 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.4 | 7.7 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.4 | 1.4 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.4 | 6.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.3 | 2.1 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) |
| 0.3 | 1.7 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.3 | 6.6 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.3 | 1.0 | GO:0010312 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.3 | 2.3 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.3 | 3.2 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.3 | 0.9 | GO:0097384 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.3 | 6.9 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.3 | 5.3 | GO:0035904 | aorta development(GO:0035904) |
| 0.3 | 1.2 | GO:1904355 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.3 | 4.9 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.3 | 2.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.3 | 4.7 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.3 | 4.1 | GO:0042026 | protein refolding(GO:0042026) |
| 0.3 | 0.8 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.3 | 4.5 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.3 | 4.5 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.3 | 2.1 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.3 | 1.3 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.3 | 6.6 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.2 | 3.5 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.2 | 1.5 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.2 | 1.2 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.2 | 1.7 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.2 | 6.1 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.2 | 3.0 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.2 | 1.9 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.2 | 4.7 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.2 | 0.9 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.2 | 1.3 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.2 | 1.3 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.2 | 2.4 | GO:0021684 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 | 4.5 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 0.8 | GO:0072314 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.2 | 0.6 | GO:0098773 | skin epidermis development(GO:0098773) |
| 0.2 | 1.2 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.2 | 2.2 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.2 | 1.2 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.2 | 1.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.2 | 3.0 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.2 | 1.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.2 | 3.9 | GO:0033555 | multicellular organismal response to stress(GO:0033555) |
| 0.2 | 3.6 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.2 | 1.5 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.2 | 2.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 3.9 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.2 | 0.9 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 | 0.6 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.1 | 1.3 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.1 | 5.0 | GO:0050727 | regulation of inflammatory response(GO:0050727) |
| 0.1 | 2.1 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 2.4 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.1 | 1.8 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.1 | 2.0 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.1 | 1.4 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.1 | 6.3 | GO:0042129 | regulation of T cell proliferation(GO:0042129) |
| 0.1 | 1.0 | GO:2000223 | regulation of BMP signaling pathway involved in heart jogging(GO:2000223) |
| 0.1 | 0.8 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 3.0 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.1 | 0.8 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 1.3 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.1 | 0.6 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.1 | 1.4 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 0.8 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.1 | 2.7 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.1 | 2.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 8.3 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.1 | 2.4 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.1 | 3.5 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 1.3 | GO:0003128 | heart field specification(GO:0003128) |
| 0.1 | 2.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 1.7 | GO:0046427 | regulation of JAK-STAT cascade(GO:0046425) positive regulation of JAK-STAT cascade(GO:0046427) regulation of STAT cascade(GO:1904892) positive regulation of STAT cascade(GO:1904894) |
| 0.1 | 0.7 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) |
| 0.1 | 0.7 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 0.8 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 1.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.3 | GO:0043363 | nucleate erythrocyte differentiation(GO:0043363) |
| 0.1 | 0.4 | GO:0032616 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.1 | 1.6 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 0.4 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.1 | 5.9 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.1 | 2.4 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 1.1 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.1 | 0.6 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 0.8 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 1.3 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 2.6 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.1 | 1.8 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 3.3 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.1 | 2.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.5 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.3 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.1 | 0.7 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.1 | 1.1 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 0.6 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.1 | 1.0 | GO:1901381 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 2.4 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 3.5 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 0.7 | GO:0010595 | positive regulation of endothelial cell migration(GO:0010595) |
| 0.1 | 1.0 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 1.6 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.1 | 2.6 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.1 | 1.1 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.1 | 0.9 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 3.1 | GO:0042493 | response to drug(GO:0042493) |
| 0.1 | 0.3 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.1 | 1.1 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.9 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.2 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.2 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 2.8 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 2.9 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 2.2 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 2.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 1.4 | GO:0031017 | exocrine pancreas development(GO:0031017) exocrine system development(GO:0035272) |
| 0.0 | 3.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.3 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 3.0 | GO:0048846 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.0 | 0.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 1.7 | GO:0008589 | regulation of smoothened signaling pathway(GO:0008589) |
| 0.0 | 1.1 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 6.2 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 1.2 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 1.0 | GO:0010906 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.9 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 2.2 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 0.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 1.0 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 3.2 | GO:0061053 | somite development(GO:0061053) |
| 0.0 | 1.9 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 2.1 | GO:0048232 | spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
| 0.0 | 0.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.3 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 6.5 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.7 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.0 | 0.6 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 1.5 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 0.2 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 2.4 | GO:0016485 | protein processing(GO:0016485) |
| 0.0 | 7.1 | GO:0001525 | angiogenesis(GO:0001525) |
| 0.0 | 0.6 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 1.0 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) cellular response to fibroblast growth factor stimulus(GO:0044344) response to fibroblast growth factor(GO:0071774) |
| 0.0 | 2.9 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.0 | 1.6 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 3.0 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 1.1 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.7 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 7.7 | GO:0010941 | regulation of cell death(GO:0010941) |
| 0.0 | 0.5 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.4 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 1.3 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.0 | 0.1 | GO:0071456 | cellular response to decreased oxygen levels(GO:0036294) cellular response to oxygen levels(GO:0071453) cellular response to hypoxia(GO:0071456) |
| 0.0 | 1.2 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 1.1 | GO:0009617 | response to bacterium(GO:0009617) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 8.8 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 1.5 | 4.4 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 1.4 | 4.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 1.0 | 2.9 | GO:0098536 | deuterosome(GO:0098536) |
| 0.9 | 4.6 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.7 | 2.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.7 | 11.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.7 | 13.6 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.5 | 1.6 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.5 | 1.9 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.5 | 8.6 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.4 | 3.9 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.4 | 4.7 | GO:0035101 | FACT complex(GO:0035101) |
| 0.4 | 2.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.3 | 4.2 | GO:0070187 | telosome(GO:0070187) |
| 0.3 | 1.1 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.2 | 12.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.2 | 1.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 0.9 | GO:1990923 | PET complex(GO:1990923) |
| 0.2 | 1.9 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 0.8 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 2.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 1.6 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 1.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 8.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 0.8 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 1.9 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 0.6 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 3.0 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 6.5 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 2.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 5.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 26.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 1.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 6.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 0.7 | GO:0035060 | brahma complex(GO:0035060) |
| 0.1 | 1.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 2.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 5.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 3.8 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.1 | 1.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.9 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 5.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 4.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 0.8 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 5.8 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 2.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.7 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 16.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 5.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 2.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 4.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 6.9 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.5 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 1.3 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 2.5 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 6.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 3.5 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 1.0 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 26.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.8 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.9 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 15.7 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 2.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.4 | GO:0070905 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 2.2 | 6.6 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 1.7 | 12.2 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 1.7 | 6.8 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 1.7 | 5.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 1.6 | 8.1 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 1.6 | 8.0 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 1.6 | 7.9 | GO:0039706 | co-receptor binding(GO:0039706) |
| 1.5 | 4.6 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 1.5 | 4.4 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 1.5 | 11.6 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 1.2 | 6.0 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 1.2 | 8.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 1.1 | 4.2 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 1.0 | 6.7 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.9 | 13.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.9 | 5.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.7 | 10.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.7 | 6.6 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.7 | 4.2 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.6 | 18.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.6 | 3.0 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.6 | 4.7 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.6 | 1.7 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.5 | 3.7 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.5 | 3.9 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.5 | 12.1 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.5 | 6.9 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.4 | 1.3 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.4 | 6.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.4 | 1.7 | GO:0071253 | connexin binding(GO:0071253) |
| 0.4 | 2.6 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.4 | 4.5 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.4 | 12.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.4 | 2.9 | GO:0035255 | GKAP/Homer scaffold activity(GO:0030160) ionotropic glutamate receptor binding(GO:0035255) |
| 0.4 | 3.9 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.4 | 2.5 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.3 | 2.6 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.3 | 2.7 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.3 | 7.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.3 | 4.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.3 | 4.2 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.3 | 1.3 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.3 | 2.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.3 | 5.5 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.2 | 5.0 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.2 | 1.9 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.2 | 1.6 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.2 | 1.6 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.2 | 3.0 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.2 | 2.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.2 | 4.5 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 0.8 | GO:0005461 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.2 | 2.8 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.2 | 2.1 | GO:0031420 | potassium ion binding(GO:0030955) alkali metal ion binding(GO:0031420) |
| 0.2 | 0.9 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.2 | 1.5 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.2 | 1.3 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.2 | 1.6 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.2 | 3.9 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.2 | 3.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 1.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 2.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 4.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 1.9 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 2.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 3.3 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.1 | 2.4 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 3.0 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 2.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 1.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.8 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 2.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 1.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 1.0 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 1.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.9 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 3.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 6.3 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 4.9 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 1.3 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 4.1 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 2.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 1.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 4.1 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 1.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 1.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 1.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 1.4 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 3.0 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.1 | 1.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 7.8 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.1 | 1.9 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 2.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.1 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 0.9 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 6.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 2.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 2.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 1.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.8 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 5.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 3.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.3 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 0.9 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 10.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 0.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 1.9 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.9 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 3.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 0.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 1.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.6 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.9 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.4 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 2.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.8 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 1.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.6 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 2.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 4.5 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 3.4 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.9 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.1 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 8.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 2.3 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.0 | 1.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 6.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 5.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 6.6 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 2.9 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 4.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 12.1 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.4 | GO:0099589 | serotonin receptor activity(GO:0099589) |
| 0.0 | 1.6 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.9 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.0 | 0.9 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 7.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.4 | GO:0016597 | amino acid binding(GO:0016597) |
| 0.0 | 0.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 1.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 9.9 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.3 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.3 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 7.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.4 | 5.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.3 | 3.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 1.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.3 | 19.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.3 | 4.7 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.3 | 7.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.3 | 9.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.2 | 15.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 2.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 3.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 1.3 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.2 | 12.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.2 | 6.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 4.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 0.4 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 1.9 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 1.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 8.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.7 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.9 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 1.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.5 | PID BCR 5PATHWAY | BCR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 11.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.6 | 13.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.5 | 4.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.4 | 8.0 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.4 | 2.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.4 | 7.1 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.4 | 5.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.3 | 3.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.3 | 3.7 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.2 | 1.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 1.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 2.4 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.2 | 1.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 6.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.2 | 2.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 10.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.2 | 2.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 1.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 3.0 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.2 | 5.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 5.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 4.4 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 2.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 2.8 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 1.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 1.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 0.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.8 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 0.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 3.6 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.1 | 1.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 1.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 1.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 3.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.9 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.2 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.6 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |