Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
Results for stat3
Z-value: 1.05
Transcription factors associated with stat3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
stat3
|
ENSDARG00000022712 | signal transducer and activator of transcription 3 (acute-phase response factor) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| stat3 | dr11_v1_chr3_+_17030665_17030704 | 0.40 | 6.1e-05 | Click! |
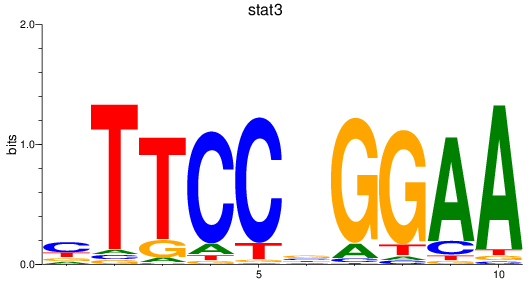
Activity profile of stat3 motif
Sorted Z-values of stat3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_10175898 | 10.61 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr24_-_33291784 | 9.90 |
ENSDART00000124938
|
si:ch1073-406l10.2
|
si:ch1073-406l10.2 |
| chr6_+_41503854 | 9.55 |
ENSDART00000136538
ENSDART00000140108 ENSDART00000084861 |
cish
|
cytokine inducible SH2-containing protein |
| chr5_+_26213874 | 9.08 |
ENSDART00000193816
ENSDART00000098514 |
oclnb
|
occludin b |
| chr21_-_27338639 | 9.00 |
ENSDART00000130632
|
hif1al2
|
hypoxia-inducible factor 1, alpha subunit, like 2 |
| chr19_+_10937932 | 8.87 |
ENSDART00000168968
|
si:ch73-347e22.8
|
si:ch73-347e22.8 |
| chr9_-_9282519 | 8.43 |
ENSDART00000146821
|
si:ch211-214p13.3
|
si:ch211-214p13.3 |
| chr12_+_17042754 | 8.16 |
ENSDART00000066439
|
ch25h
|
cholesterol 25-hydroxylase |
| chr1_-_51734524 | 8.10 |
ENSDART00000109640
ENSDART00000122628 |
junba
|
JunB proto-oncogene, AP-1 transcription factor subunit a |
| chr3_-_57425961 | 7.61 |
ENSDART00000033716
|
socs3a
|
suppressor of cytokine signaling 3a |
| chr14_-_30747686 | 7.39 |
ENSDART00000008373
|
fosl1a
|
FOS-like antigen 1a |
| chr11_-_6265574 | 7.33 |
ENSDART00000181974
ENSDART00000104405 |
ccl25b
|
chemokine (C-C motif) ligand 25b |
| chr21_+_45626136 | 7.26 |
ENSDART00000158742
|
irf1b
|
interferon regulatory factor 1b |
| chr21_+_43328685 | 7.12 |
ENSDART00000109620
ENSDART00000139668 |
sept8a
|
septin 8a |
| chr17_-_10838434 | 7.00 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr21_-_35419486 | 6.94 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr3_-_44059902 | 6.90 |
ENSDART00000158485
ENSDART00000159088 ENSDART00000165628 |
il4r.1
|
interleukin 4 receptor, tandem duplicate 1 |
| chr25_-_21822426 | 6.56 |
ENSDART00000151993
|
zgc:158222
|
zgc:158222 |
| chr22_+_22004082 | 6.52 |
ENSDART00000148375
|
gna15.2
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 2 |
| chr14_+_11457500 | 6.27 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr6_+_41099787 | 6.15 |
ENSDART00000186884
|
fkbp5
|
FK506 binding protein 5 |
| chr3_-_39488482 | 6.10 |
ENSDART00000135192
|
zgc:100868
|
zgc:100868 |
| chr3_-_4253214 | 5.93 |
ENSDART00000155807
|
si:dkey-175d9.2
|
si:dkey-175d9.2 |
| chr3_+_26813058 | 5.86 |
ENSDART00000055537
|
socs1a
|
suppressor of cytokine signaling 1a |
| chr5_+_28271412 | 5.56 |
ENSDART00000031727
|
vamp8
|
vesicle-associated membrane protein 8 (endobrevin) |
| chr7_+_26167420 | 5.39 |
ENSDART00000173941
|
si:ch211-196f2.6
|
si:ch211-196f2.6 |
| chr11_-_17964525 | 5.25 |
ENSDART00000018948
|
cishb
|
cytokine inducible SH2-containing protein b |
| chr16_-_38001040 | 5.17 |
ENSDART00000133861
ENSDART00000138711 ENSDART00000143846 ENSDART00000146564 |
si:ch211-198c19.3
|
si:ch211-198c19.3 |
| chr14_+_11395383 | 5.04 |
ENSDART00000106657
|
glod5
|
glyoxalase domain containing 5 |
| chr19_+_43359075 | 4.96 |
ENSDART00000148287
ENSDART00000149856 ENSDART00000188236 ENSDART00000136695 ENSDART00000193859 |
yrk
|
Yes-related kinase |
| chr5_-_12587053 | 4.93 |
ENSDART00000162780
|
vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr20_-_19858936 | 4.85 |
ENSDART00000161579
|
ptk2bb
|
protein tyrosine kinase 2 beta, b |
| chr6_-_34220641 | 4.75 |
ENSDART00000102391
|
dmrt2b
|
doublesex and mab-3 related transcription factor 2b |
| chr4_+_73215536 | 4.70 |
ENSDART00000174290
|
LO018260.2
|
Danio rerio protein NLRC3-like (LOC101883187), mRNA. |
| chr23_-_5101847 | 4.70 |
ENSDART00000122240
|
etv7
|
ets variant 7 |
| chr16_-_22930925 | 4.60 |
ENSDART00000133819
|
si:dkey-246i14.3
|
si:dkey-246i14.3 |
| chr17_+_16046314 | 4.52 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr10_+_13209580 | 4.48 |
ENSDART00000000887
ENSDART00000136932 |
rassf6
|
Ras association (RalGDS/AF-6) domain family 6 |
| chr4_+_7391400 | 4.42 |
ENSDART00000169111
ENSDART00000186395 |
tnni4a
|
troponin I4a |
| chr4_+_10616626 | 4.42 |
ENSDART00000067251
ENSDART00000143690 |
cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr24_-_27452488 | 4.40 |
ENSDART00000136433
|
ccl34b.8
|
chemokine (C-C motif) ligand 34b, duplicate 8 |
| chr4_+_76735113 | 4.39 |
ENSDART00000075602
|
ms4a17a.6
|
membrane-spanning 4-domains, subfamily A, member 17A.6 |
| chr22_-_15578402 | 4.28 |
ENSDART00000062986
|
hsh2d
|
hematopoietic SH2 domain containing |
| chr3_-_44012748 | 4.27 |
ENSDART00000167248
ENSDART00000157463 ENSDART00000159111 |
il4r.1
|
interleukin 4 receptor, tandem duplicate 1 |
| chr23_-_5783421 | 4.26 |
ENSDART00000131521
ENSDART00000019455 |
csrp1a
|
cysteine and glycine-rich protein 1a |
| chr9_-_31135901 | 4.22 |
ENSDART00000113027
ENSDART00000193210 ENSDART00000128896 |
si:ch211-184m13.4
|
si:ch211-184m13.4 |
| chr9_-_23922011 | 4.15 |
ENSDART00000145734
|
col6a3
|
collagen, type VI, alpha 3 |
| chr7_-_22790630 | 4.06 |
ENSDART00000173496
|
si:ch211-15b10.6
|
si:ch211-15b10.6 |
| chr8_-_23240156 | 4.02 |
ENSDART00000131632
|
ptk6a
|
PTK6 protein tyrosine kinase 6a |
| chr13_+_39532050 | 3.96 |
ENSDART00000019379
|
marveld1
|
MARVEL domain containing 1 |
| chr21_-_43328056 | 3.94 |
ENSDART00000114955
|
sowahaa
|
sosondowah ankyrin repeat domain family member Aa |
| chr12_-_7234915 | 3.91 |
ENSDART00000048866
|
ipmkb
|
inositol polyphosphate multikinase b |
| chr5_-_23619911 | 3.91 |
ENSDART00000020004
|
mtmr2
|
myotubularin related protein 2 |
| chr16_+_38394371 | 3.90 |
ENSDART00000137954
|
cd83
|
CD83 molecule |
| chr9_-_23922778 | 3.83 |
ENSDART00000135769
|
col6a3
|
collagen, type VI, alpha 3 |
| chr16_+_42772678 | 3.71 |
ENSDART00000155575
|
si:ch211-135n15.2
|
si:ch211-135n15.2 |
| chr21_+_13387965 | 3.68 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr4_+_68922112 | 3.59 |
ENSDART00000159074
|
si:dkey-264f17.2
|
si:dkey-264f17.2 |
| chr20_+_18993452 | 3.58 |
ENSDART00000025509
|
tdh
|
L-threonine dehydrogenase |
| chr22_-_5323482 | 3.58 |
ENSDART00000145785
|
s1pr4
|
sphingosine-1-phosphate receptor 4 |
| chr8_+_23152244 | 3.53 |
ENSDART00000036801
ENSDART00000184298 |
slc17a9a
|
solute carrier family 17 (vesicular nucleotide transporter), member 9a |
| chr17_+_16046132 | 3.48 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr20_+_18994059 | 3.40 |
ENSDART00000152380
|
tdh
|
L-threonine dehydrogenase |
| chr3_-_8130926 | 3.35 |
ENSDART00000189495
|
si:ch211-51i16.1
|
si:ch211-51i16.1 |
| chr13_-_37642890 | 3.27 |
ENSDART00000146483
ENSDART00000136071 ENSDART00000111786 |
si:dkey-188i13.9
|
si:dkey-188i13.9 |
| chr19_+_10587307 | 3.26 |
ENSDART00000131225
|
si:dkey-211g8.4
|
si:dkey-211g8.4 |
| chr3_-_32275975 | 3.07 |
ENSDART00000178448
|
cpt1cb
|
carnitine palmitoyltransferase 1Cb |
| chr5_+_25317061 | 3.06 |
ENSDART00000170097
|
trpm6
|
transient receptor potential cation channel, subfamily M, member 6 |
| chr12_-_19346678 | 3.05 |
ENSDART00000044860
|
maff
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr7_-_22941472 | 3.05 |
ENSDART00000190334
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr9_-_14084743 | 3.03 |
ENSDART00000056105
|
fer1l6
|
fer-1-like family member 6 |
| chr4_-_4592287 | 3.02 |
ENSDART00000155287
|
rassf3
|
Ras association (RalGDS/AF-6) domain family member 3 |
| chr3_+_472158 | 2.97 |
ENSDART00000134971
|
zgc:194659
|
zgc:194659 |
| chr21_-_43457554 | 2.97 |
ENSDART00000085039
|
stk26
|
serine/threonine protein kinase 26 |
| chr4_+_72668095 | 2.94 |
ENSDART00000182282
ENSDART00000162637 |
CABZ01054391.1
|
|
| chr18_-_14860435 | 2.89 |
ENSDART00000018502
|
mapk12a
|
mitogen-activated protein kinase 12a |
| chr4_-_12323228 | 2.88 |
ENSDART00000081089
|
il17ra1a
|
interleukin 17 receptor A1a |
| chr20_-_37813863 | 2.86 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr6_+_36821621 | 2.85 |
ENSDART00000104157
|
tmem45a
|
transmembrane protein 45a |
| chr14_-_6987649 | 2.84 |
ENSDART00000060990
|
eif4ebp3l
|
eukaryotic translation initiation factor 4E binding protein 3, like |
| chr3_+_16922226 | 2.84 |
ENSDART00000017646
|
atp6v0a1a
|
ATPase H+ transporting V0 subunit a1a |
| chr21_-_21781158 | 2.79 |
ENSDART00000113734
|
chrdl2
|
chordin-like 2 |
| chr20_-_35841570 | 2.78 |
ENSDART00000037855
|
tnfrsf21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr16_-_27749172 | 2.78 |
ENSDART00000145198
|
steap4
|
STEAP family member 4 |
| chr6_+_11989537 | 2.76 |
ENSDART00000190817
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr22_-_13165186 | 2.74 |
ENSDART00000105762
|
ahr2
|
aryl hydrocarbon receptor 2 |
| chr20_+_35433192 | 2.61 |
ENSDART00000017485
|
sf3b6
|
splicing factor 3b, subunit 6 |
| chr17_+_25397070 | 2.57 |
ENSDART00000164254
|
zgc:154055
|
zgc:154055 |
| chr15_-_3282220 | 2.57 |
ENSDART00000092942
|
slc25a15a
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15a |
| chr2_+_20967673 | 2.50 |
ENSDART00000057174
|
arpc5a
|
actin related protein 2/3 complex, subunit 5A |
| chr18_-_16801033 | 2.49 |
ENSDART00000100100
|
admb
|
adrenomedullin b |
| chr20_+_25552057 | 2.47 |
ENSDART00000102913
|
cyp2v1
|
cytochrome P450, family 2, subfamily V, polypeptide 1 |
| chr8_+_52515188 | 2.41 |
ENSDART00000163668
|
si:ch1073-392o20.2
|
si:ch1073-392o20.2 |
| chr2_+_4402765 | 2.36 |
ENSDART00000159525
|
bambib
|
BMP and activin membrane-bound inhibitor homolog (Xenopus laevis) b |
| chr11_+_16040517 | 2.36 |
ENSDART00000111284
|
agtrap
|
angiotensin II receptor-associated protein |
| chr2_+_26288301 | 2.35 |
ENSDART00000017668
|
ptbp1a
|
polypyrimidine tract binding protein 1a |
| chr3_+_59899452 | 2.34 |
ENSDART00000064311
|
arhgdia
|
Rho GDP dissociation inhibitor (GDI) alpha |
| chr7_-_28696556 | 2.33 |
ENSDART00000148822
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr7_+_28901786 | 2.31 |
ENSDART00000113998
|
dok4
|
docking protein 4 |
| chr6_-_55958705 | 2.30 |
ENSDART00000155963
|
eya2
|
EYA transcriptional coactivator and phosphatase 2 |
| chr1_-_9114936 | 2.20 |
ENSDART00000163624
ENSDART00000132910 |
si:ch211-14k19.8
vap
|
si:ch211-14k19.8 vascular associated protein |
| chr15_+_23550890 | 2.18 |
ENSDART00000009796
ENSDART00000152720 |
MARK4
|
si:dkey-31m14.7 |
| chr9_+_1313418 | 2.18 |
ENSDART00000191361
|
casp8l2
|
caspase 8, apoptosis-related cysteine peptidase, like 2 |
| chr20_-_39273987 | 2.17 |
ENSDART00000127173
|
clu
|
clusterin |
| chr11_+_12720171 | 2.14 |
ENSDART00000135761
ENSDART00000122812 |
ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr8_+_19548985 | 2.13 |
ENSDART00000123104
|
notch2
|
notch 2 |
| chr19_+_23298037 | 2.13 |
ENSDART00000183948
|
irgf1
|
immunity-related GTPase family, f1 |
| chr16_+_54513267 | 2.11 |
ENSDART00000161923
|
CABZ01088508.1
|
|
| chr21_+_35215810 | 2.10 |
ENSDART00000135256
|
ubtd2
|
ubiquitin domain containing 2 |
| chr14_+_15257658 | 2.09 |
ENSDART00000161625
ENSDART00000193577 |
si:dkey-77g12.4
si:dkey-203a12.5
|
si:dkey-77g12.4 si:dkey-203a12.5 |
| chr16_+_25809235 | 2.09 |
ENSDART00000181196
|
irgq1
|
immunity-related GTPase family, q1 |
| chr20_-_25412181 | 2.05 |
ENSDART00000062780
|
itsn2a
|
intersectin 2a |
| chr11_+_4026229 | 2.01 |
ENSDART00000041417
|
camk1b
|
calcium/calmodulin-dependent protein kinase Ib |
| chr21_-_5879897 | 2.01 |
ENSDART00000184034
|
rpl35
|
ribosomal protein L35 |
| chr7_-_24373662 | 1.95 |
ENSDART00000173865
|
PTGR1 (1 of many)
|
si:dkey-11k2.7 |
| chr25_+_15933411 | 1.93 |
ENSDART00000191581
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr5_-_26466169 | 1.92 |
ENSDART00000144035
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr24_+_31361407 | 1.92 |
ENSDART00000162668
|
cremb
|
cAMP responsive element modulator b |
| chr15_+_16521785 | 1.91 |
ENSDART00000062191
|
galnt17
|
polypeptide N-acetylgalactosaminyltransferase 17 |
| chr4_+_76637548 | 1.90 |
ENSDART00000133799
|
ms4a17a.9
|
membrane-spanning 4-domains, subfamily A, member 17A.9 |
| chr1_-_56596701 | 1.90 |
ENSDART00000133693
|
LO018605.1
|
|
| chr20_+_46620374 | 1.86 |
ENSDART00000005548
|
batf
|
basic leucine zipper transcription factor, ATF-like |
| chr1_-_10669436 | 1.85 |
ENSDART00000152421
|
si:dkey-31e10.5
|
si:dkey-31e10.5 |
| chr7_+_12950507 | 1.84 |
ENSDART00000067629
ENSDART00000158004 |
saa
|
serum amyloid A |
| chr19_+_8973042 | 1.80 |
ENSDART00000039597
|
crabp2b
|
cellular retinoic acid binding protein 2, b |
| chr1_-_9195629 | 1.73 |
ENSDART00000143587
ENSDART00000192174 |
ern2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr2_+_41526904 | 1.73 |
ENSDART00000127520
|
acvr1l
|
activin A receptor, type 1 like |
| chr23_-_33558161 | 1.73 |
ENSDART00000018301
|
itga5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr9_-_43644261 | 1.72 |
ENSDART00000023684
|
cwc22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr8_+_8671229 | 1.72 |
ENSDART00000131963
|
usp11
|
ubiquitin specific peptidase 11 |
| chr19_+_7916790 | 1.70 |
ENSDART00000081584
|
si:dkey-266f7.1
|
si:dkey-266f7.1 |
| chr6_+_37655078 | 1.70 |
ENSDART00000122199
ENSDART00000065127 |
cyfip1
|
cytoplasmic FMR1 interacting protein 1 |
| chr5_+_20103085 | 1.67 |
ENSDART00000079424
|
si:rp71-1c23.3
|
si:rp71-1c23.3 |
| chr24_-_26383978 | 1.66 |
ENSDART00000031426
|
skilb
|
SKI-like proto-oncogene b |
| chr12_+_22657925 | 1.65 |
ENSDART00000153048
|
si:dkey-219e21.4
|
si:dkey-219e21.4 |
| chr14_-_32744464 | 1.65 |
ENSDART00000075617
|
sox3
|
SRY (sex determining region Y)-box 3 |
| chr3_+_44062576 | 1.59 |
ENSDART00000161277
ENSDART00000168784 |
nsmce1
|
NSE1 homolog, SMC5-SMC6 complex component |
| chr24_+_37406535 | 1.59 |
ENSDART00000138264
|
si:ch211-183d21.1
|
si:ch211-183d21.1 |
| chr12_+_38983989 | 1.59 |
ENSDART00000156673
|
si:ch73-181m17.1
|
si:ch73-181m17.1 |
| chr9_-_12659140 | 1.58 |
ENSDART00000058565
|
pttg1ipb
|
PTTG1 interacting protein b |
| chr21_-_34844316 | 1.58 |
ENSDART00000029708
|
zgc:56585
|
zgc:56585 |
| chr18_+_14684115 | 1.57 |
ENSDART00000108469
|
spata2l
|
spermatogenesis associated 2-like |
| chr6_+_13232934 | 1.55 |
ENSDART00000089725
|
ino80db
|
INO80 complex subunit Db |
| chr21_-_22676323 | 1.53 |
ENSDART00000167392
|
gig2h
|
grass carp reovirus (GCRV)-induced gene 2h |
| chr9_+_26103814 | 1.51 |
ENSDART00000026011
|
efnb2a
|
ephrin-B2a |
| chr9_+_40825065 | 1.50 |
ENSDART00000137673
|
si:dkey-95p16.2
|
si:dkey-95p16.2 |
| chr7_+_41313568 | 1.50 |
ENSDART00000016660
|
zgc:165532
|
zgc:165532 |
| chr24_-_27461295 | 1.49 |
ENSDART00000110748
|
ccl34b.9
|
chemokine (C-C motif) ligand 34b, duplicate 9 |
| chr5_-_42878178 | 1.48 |
ENSDART00000162981
|
CXCL11 (1 of many)
|
C-X-C motif chemokine ligand 11 |
| chr8_-_49766205 | 1.48 |
ENSDART00000137941
ENSDART00000097919 ENSDART00000147309 |
hnrnpk
|
heterogeneous nuclear ribonucleoprotein K |
| chr4_+_3455665 | 1.47 |
ENSDART00000058277
|
znf800b
|
zinc finger protein 800b |
| chr10_-_4375190 | 1.46 |
ENSDART00000016102
|
CABZ01073795.1
|
|
| chr8_+_26329482 | 1.46 |
ENSDART00000048676
|
prkar2aa
|
protein kinase, cAMP-dependent, regulatory, type II, alpha A |
| chr11_+_16040680 | 1.45 |
ENSDART00000157703
|
agtrap
|
angiotensin II receptor-associated protein |
| chr4_+_49322310 | 1.42 |
ENSDART00000184154
ENSDART00000167162 |
BX942819.1
|
|
| chr9_+_13985567 | 1.41 |
ENSDART00000102296
|
cd28
|
CD28 molecule |
| chr4_+_57580303 | 1.41 |
ENSDART00000166492
ENSDART00000103025 ENSDART00000170786 |
il17ra1b
il17ra2
|
interleukin 17 receptor A1b interleukin 17 receptor A2 |
| chr23_-_19715799 | 1.40 |
ENSDART00000142072
ENSDART00000032744 ENSDART00000131860 |
rpl10
|
ribosomal protein L10 |
| chr23_+_5524247 | 1.36 |
ENSDART00000189679
ENSDART00000083622 |
tead3a
|
TEA domain family member 3 a |
| chr19_-_35052451 | 1.33 |
ENSDART00000152030
|
macf1a
|
microtubule-actin crosslinking factor 1a |
| chr21_-_34844059 | 1.33 |
ENSDART00000136402
|
zgc:56585
|
zgc:56585 |
| chr3_+_58984463 | 1.32 |
ENSDART00000162097
|
CABZ01044156.1
|
|
| chr3_-_7912661 | 1.31 |
ENSDART00000161400
|
trim35-21
|
tripartite motif containing 35-21 |
| chr22_-_12890921 | 1.30 |
ENSDART00000146484
|
nab1a
|
NGFI-A binding protein 1a (EGR1 binding protein 1) |
| chr16_+_31921812 | 1.30 |
ENSDART00000176928
ENSDART00000193733 |
rps9
|
ribosomal protein S9 |
| chr18_-_26811959 | 1.28 |
ENSDART00000086250
|
znf592
|
zinc finger protein 592 |
| chr24_-_39858710 | 1.28 |
ENSDART00000134251
|
slc12a7b
|
solute carrier family 12 (potassium/chloride transporter), member 7b |
| chr9_-_54304684 | 1.27 |
ENSDART00000109512
|
il13
|
interleukin 13 |
| chr1_+_55600504 | 1.27 |
ENSDART00000123946
|
umod
|
uromodulin |
| chr3_-_32170850 | 1.26 |
ENSDART00000055307
ENSDART00000157366 |
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr22_+_5851122 | 1.26 |
ENSDART00000082044
|
zmp:0000001161
|
zmp:0000001161 |
| chr2_-_37837056 | 1.25 |
ENSDART00000158179
ENSDART00000160317 ENSDART00000171409 |
mettl17
|
methyltransferase like 17 |
| chr5_-_30620625 | 1.25 |
ENSDART00000098273
|
tcnl
|
transcobalamin like |
| chr24_-_37877743 | 1.24 |
ENSDART00000105658
|
tmem204
|
transmembrane protein 204 |
| chr8_-_31701157 | 1.24 |
ENSDART00000141799
|
fbxo4
|
F-box protein 4 |
| chr16_-_20870143 | 1.23 |
ENSDART00000169541
ENSDART00000040727 |
tax1bp1b
|
Tax1 (human T-cell leukemia virus type I) binding protein 1b |
| chr16_+_31922065 | 1.22 |
ENSDART00000131661
ENSDART00000144194 ENSDART00000145510 |
rps9
|
ribosomal protein S9 |
| chr17_+_33415319 | 1.21 |
ENSDART00000140805
ENSDART00000025501 ENSDART00000146447 |
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr11_+_5681762 | 1.19 |
ENSDART00000179139
|
arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr3_+_1219344 | 1.16 |
ENSDART00000161945
|
rrp7a
|
ribosomal RNA processing 7 homolog A |
| chr7_+_25053331 | 1.16 |
ENSDART00000173998
|
si:dkey-23i12.7
|
si:dkey-23i12.7 |
| chr8_-_25707421 | 1.15 |
ENSDART00000184404
|
sema3ga
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ga |
| chr20_+_53521648 | 1.13 |
ENSDART00000139794
|
pak6b
|
p21 protein (Cdc42/Rac)-activated kinase 6b |
| chr20_-_5400395 | 1.13 |
ENSDART00000168103
|
snw1
|
SNW domain containing 1 |
| chr7_+_32369026 | 1.12 |
ENSDART00000169588
|
lgr4
|
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr19_-_30420602 | 1.12 |
ENSDART00000144121
|
ppp1r10
|
protein phosphatase 1, regulatory subunit 10 |
| chr10_-_5568379 | 1.11 |
ENSDART00000164558
ENSDART00000005292 |
syk
|
spleen tyrosine kinase |
| chr11_-_22303678 | 1.11 |
ENSDART00000159527
ENSDART00000159681 |
tfeb
|
transcription factor EB |
| chr7_+_17953589 | 1.09 |
ENSDART00000174778
ENSDART00000113120 |
taf6l
|
TAF6-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor |
| chr9_-_8670158 | 1.09 |
ENSDART00000077296
|
col4a1
|
collagen, type IV, alpha 1 |
| chr16_-_45398408 | 1.08 |
ENSDART00000004052
|
rgl2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr6_-_2454691 | 1.06 |
ENSDART00000192820
|
MED8
|
mediator complex subunit 8 |
| chr16_+_10841163 | 1.05 |
ENSDART00000065467
|
dedd1
|
death effector domain-containing 1 |
| chr15_-_1485086 | 1.04 |
ENSDART00000191651
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr7_+_48319916 | 1.03 |
ENSDART00000052122
|
crabp1b
|
cellular retinoic acid binding protein 1b |
| chr20_-_20932760 | 1.02 |
ENSDART00000152415
ENSDART00000039907 |
btbd6b
|
BTB (POZ) domain containing 6b |
| chr9_-_46072805 | 1.02 |
ENSDART00000169682
|
hdac4
|
histone deacetylase 4 |
| chr25_-_21085661 | 1.01 |
ENSDART00000099355
|
prr5a
|
proline rich 5a (renal) |
| chr4_+_57194439 | 1.00 |
ENSDART00000075265
|
si:dkey-16p6.4
|
si:dkey-16p6.4 |
| chr3_-_40933415 | 0.98 |
ENSDART00000055201
|
foxk1
|
forkhead box K1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of stat3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 11.2 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 2.4 | 7.3 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 1.9 | 13.4 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 1.8 | 5.4 | GO:0002418 | response to tumor cell(GO:0002347) natural killer cell cytokine production(GO:0002370) immune response to tumor cell(GO:0002418) natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of response to biotic stimulus(GO:0002833) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 1.4 | 7.0 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 1.3 | 3.9 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) |
| 1.0 | 7.6 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.9 | 2.8 | GO:0031642 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.9 | 5.6 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.9 | 7.0 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.9 | 2.6 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.7 | 2.8 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.7 | 2.8 | GO:0015677 | copper ion import(GO:0015677) |
| 0.6 | 2.5 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.6 | 7.3 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.6 | 2.4 | GO:0060467 | negative regulation of fertilization(GO:0060467) prevention of polyspermy(GO:0060468) cortical granule exocytosis(GO:0060471) |
| 0.6 | 1.1 | GO:0033005 | positive regulation of mast cell activation(GO:0033005) |
| 0.6 | 9.9 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.5 | 1.4 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.5 | 1.8 | GO:0001909 | leukocyte mediated cytotoxicity(GO:0001909) |
| 0.4 | 3.1 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.4 | 1.7 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.4 | 1.7 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.4 | 2.3 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.4 | 9.0 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.4 | 4.9 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.3 | 1.7 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.3 | 14.0 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.3 | 3.0 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.3 | 3.3 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.3 | 1.5 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.3 | 1.7 | GO:0090245 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) axis elongation involved in somitogenesis(GO:0090245) |
| 0.3 | 0.8 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.3 | 2.3 | GO:2001239 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.3 | 1.0 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.3 | 1.5 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.2 | 2.7 | GO:0001541 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) |
| 0.2 | 8.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.2 | 2.8 | GO:0001845 | phagolysosome assembly(GO:0001845) |
| 0.2 | 5.0 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.2 | 0.9 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.2 | 1.3 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.2 | 1.0 | GO:0016115 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.2 | 6.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 3.9 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.2 | 1.2 | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.2 | 1.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.2 | 4.8 | GO:0007548 | sex differentiation(GO:0007548) |
| 0.2 | 0.5 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.2 | 0.5 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.2 | 4.0 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 0.4 | GO:0003091 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 2.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 1.8 | GO:0048385 | regulation of retinoic acid receptor signaling pathway(GO:0048385) |
| 0.1 | 6.7 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 2.0 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 3.6 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.1 | 0.6 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 1.0 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.1 | 4.3 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.1 | 10.6 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.1 | 8.0 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 4.4 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 6.9 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.1 | 1.0 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 1.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 1.0 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.3 | GO:0035521 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 3.3 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.1 | 4.7 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 1.6 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.1 | 0.7 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 0.2 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.1 | 0.5 | GO:0032366 | intracellular sterol transport(GO:0032366) |
| 0.1 | 4.0 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.1 | 6.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 0.6 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.4 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.7 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.1 | 1.1 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 4.0 | GO:0042552 | myelination(GO:0042552) |
| 0.1 | 3.1 | GO:0051262 | protein tetramerization(GO:0051262) |
| 0.1 | 0.2 | GO:1904353 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.1 | 1.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 1.3 | GO:0071219 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.1 | 1.6 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.1 | 2.2 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 1.0 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.1 | 0.8 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 1.1 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 3.8 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.0 | 1.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 1.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.5 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 4.1 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 1.6 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 2.3 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 1.4 | GO:0042129 | regulation of T cell proliferation(GO:0042129) |
| 0.0 | 1.2 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 2.3 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.5 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.0 | 0.7 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 3.1 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.0 | 1.5 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.8 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.7 | GO:1904018 | positive regulation of angiogenesis(GO:0045766) positive regulation of vasculature development(GO:1904018) |
| 0.0 | 0.6 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 1.1 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 1.9 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.4 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.0 | 1.4 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.5 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.0 | 1.1 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of chemotaxis(GO:0050922) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 6.2 | GO:0000398 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.7 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 1.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.9 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 1.7 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 1.4 | GO:0048738 | cardiac muscle tissue development(GO:0048738) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 10.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.9 | 7.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.6 | 1.7 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.4 | 25.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.4 | 1.2 | GO:0034456 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.3 | 1.7 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.3 | 1.7 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.3 | 2.8 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.3 | 1.1 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.2 | 1.6 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.2 | 2.7 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 7.1 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.2 | 1.3 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.2 | 1.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 1.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.9 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 0.4 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 2.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.7 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 5.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 2.6 | GO:0005689 | U2 snRNP(GO:0005686) U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 14.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 1.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 9.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.9 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.6 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.1 | 2.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 0.7 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 6.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 3.9 | GO:0019814 | immunoglobulin complex(GO:0019814) |
| 0.1 | 0.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.5 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 3.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 3.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 2.5 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.9 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 1.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 4.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.5 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 2.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.1 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 1.7 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 11.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.3 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 7.8 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 3.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.5 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 2.6 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 3.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 2.7 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.3 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.5 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 6.1 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 1.4 | 7.0 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 1.4 | 7.0 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 1.3 | 3.9 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 1.2 | 8.2 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.8 | 25.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.7 | 2.8 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.6 | 2.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.4 | 3.6 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.4 | 2.8 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.4 | 4.7 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.4 | 3.9 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.3 | 5.9 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.3 | 3.3 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.3 | 2.6 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.3 | 4.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.3 | 7.3 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.2 | 1.5 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.2 | 2.8 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.2 | 14.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.2 | 0.7 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.2 | 4.3 | GO:0042805 | actinin binding(GO:0042805) |
| 0.2 | 2.8 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.1 | 1.7 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.4 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 1.0 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 1.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.7 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 0.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 11.2 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.1 | 1.7 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 6.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 3.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 2.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.0 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 1.0 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 1.7 | GO:0004407 | histone deacetylase activity(GO:0004407) NAD-dependent histone deacetylase activity(GO:0017136) |
| 0.1 | 2.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 0.2 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.1 | 1.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.4 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.1 | 1.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 2.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.5 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 1.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.1 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 9.2 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.1 | 1.3 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 2.5 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 2.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 8.6 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 2.3 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0102345 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 1.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.7 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 1.7 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 1.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 5.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 4.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 2.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 2.7 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.3 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 1.7 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 7.2 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 2.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.8 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 4.2 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 23.0 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 9.6 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.3 | 3.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.3 | 2.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 18.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.2 | 9.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 1.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 1.0 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 1.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 2.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 2.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 2.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 2.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 2.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 2.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 8.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.4 | 7.6 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.3 | 3.9 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.3 | 2.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.3 | 7.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.2 | 9.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 1.4 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 0.7 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 2.7 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 3.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 0.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 6.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 3.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 3.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.9 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.6 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |