Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
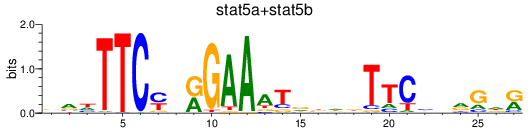
Results for stat5a+stat5b
Z-value: 0.63
Transcription factors associated with stat5a+stat5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
stat5a
|
ENSDARG00000019392 | signal transducer and activator of transcription 5a |
|
stat5b
|
ENSDARG00000055588 | signal transducer and activator of transcription 5b |
|
stat5a
|
ENSDARG00000113871 | signal transducer and activator of transcription 5a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| stat5b | dr11_v1_chr12_-_13650344_13650472 | -0.39 | 8.9e-05 | Click! |
| stat5a | dr11_v1_chr3_+_17314997_17314997 | 0.11 | 2.9e-01 | Click! |
Activity profile of stat5a+stat5b motif
Sorted Z-values of stat5a+stat5b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_37933983 | 4.79 |
ENSDART00000185364
|
scn4bb
|
sodium channel, voltage-gated, type IV, beta b |
| chr23_+_26017227 | 3.51 |
ENSDART00000002939
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr19_-_32804535 | 3.51 |
ENSDART00000175613
ENSDART00000052098 |
nt5c1aa
|
5'-nucleotidase, cytosolic IAa |
| chr19_-_12648408 | 3.35 |
ENSDART00000103692
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr25_+_6186823 | 3.23 |
ENSDART00000153526
|
oaz2a
|
ornithine decarboxylase antizyme 2a |
| chr2_+_22659787 | 3.06 |
ENSDART00000043956
|
zgc:161973
|
zgc:161973 |
| chr22_+_36914636 | 2.91 |
ENSDART00000150948
|
pimr205
|
Pim proto-oncogene, serine/threonine kinase, related 205 |
| chr8_-_42624454 | 2.74 |
ENSDART00000075550
|
kazald3
|
Kazal-type serine peptidase inhibitor domain 3 |
| chr21_-_38619305 | 2.54 |
ENSDART00000139178
|
slc25a14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr2_+_37140448 | 2.51 |
ENSDART00000045016
ENSDART00000142940 |
pex19
|
peroxisomal biogenesis factor 19 |
| chr15_-_15449929 | 2.38 |
ENSDART00000101918
|
proca1
|
protein interacting with cyclin A1 |
| chr12_-_314899 | 2.28 |
ENSDART00000066579
|
pts
|
6-pyruvoyltetrahydropterin synthase |
| chr20_+_10166297 | 2.26 |
ENSDART00000141877
|
kcnk10a
|
potassium channel, subfamily K, member 10a |
| chr2_-_51794472 | 2.25 |
ENSDART00000186652
|
BX908782.3
|
|
| chr22_+_7480465 | 2.15 |
ENSDART00000034545
|
CELA1 (1 of many)
|
zgc:92745 |
| chr16_+_34523515 | 2.10 |
ENSDART00000041007
|
stmn1b
|
stathmin 1b |
| chr15_-_32383529 | 2.02 |
ENSDART00000028349
|
c4
|
complement component 4 |
| chr15_-_2857961 | 2.01 |
ENSDART00000033263
|
ankrd49
|
ankyrin repeat domain 49 |
| chr2_-_49978227 | 2.00 |
ENSDART00000142835
|
hecw1b
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1b |
| chr25_-_27842654 | 1.98 |
ENSDART00000154852
ENSDART00000156906 |
asb15a
|
ankyrin repeat and SOCS box containing 15a |
| chr7_-_36358735 | 1.95 |
ENSDART00000188392
|
fto
|
fat mass and obesity associated |
| chr3_-_46818001 | 1.81 |
ENSDART00000166505
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr20_+_48754045 | 1.80 |
ENSDART00000062578
|
zgc:110348
|
zgc:110348 |
| chr7_-_36358303 | 1.80 |
ENSDART00000130028
|
fto
|
fat mass and obesity associated |
| chr5_+_27137473 | 1.80 |
ENSDART00000181833
|
unc5db
|
unc-5 netrin receptor Db |
| chr12_+_22680115 | 1.78 |
ENSDART00000152879
|
ablim2
|
actin binding LIM protein family, member 2 |
| chr15_-_32383340 | 1.78 |
ENSDART00000185632
|
c4
|
complement component 4 |
| chr4_+_11479705 | 1.76 |
ENSDART00000019458
ENSDART00000150587 ENSDART00000139370 ENSDART00000135826 |
asb13a.1
|
ankyrin repeat and SOCS box containing 13a, tandem duplicate 1 |
| chr7_+_58751504 | 1.74 |
ENSDART00000024185
|
zgc:56231
|
zgc:56231 |
| chr3_-_46817838 | 1.70 |
ENSDART00000028610
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr18_+_6479963 | 1.70 |
ENSDART00000092752
ENSDART00000136333 |
wash1
|
WAS protein family homolog 1 |
| chr12_-_46959990 | 1.65 |
ENSDART00000084557
|
lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr10_-_43568239 | 1.64 |
ENSDART00000131731
ENSDART00000097433 ENSDART00000131309 |
mef2ca
|
myocyte enhancer factor 2ca |
| chr2_+_7192966 | 1.62 |
ENSDART00000142735
|
si:ch211-13f8.1
|
si:ch211-13f8.1 |
| chr19_+_25504645 | 1.59 |
ENSDART00000143292
|
col28a1a
|
collagen, type XXVIII, alpha 1a |
| chr20_+_46897504 | 1.58 |
ENSDART00000158124
|
si:ch73-21k16.1
|
si:ch73-21k16.1 |
| chr7_-_11605185 | 1.54 |
ENSDART00000169291
ENSDART00000113904 |
stard5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr3_+_26905024 | 1.53 |
ENSDART00000077835
|
dexi
|
Dexi homolog (mouse) |
| chr6_+_16870004 | 1.51 |
ENSDART00000154794
|
pimr21
|
Pim proto-oncogene, serine/threonine kinase, related 21 |
| chr1_-_7582859 | 1.51 |
ENSDART00000110696
|
mxb
|
myxovirus (influenza) resistance B |
| chr20_+_17581881 | 1.49 |
ENSDART00000182832
|
cdh2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr10_-_24868581 | 1.49 |
ENSDART00000193055
|
CABZ01058107.1
|
|
| chr13_-_49819027 | 1.45 |
ENSDART00000067824
|
b3galnt2
|
beta-1,3-N-acetylgalactosaminyltransferase 2 |
| chr18_+_41772474 | 1.45 |
ENSDART00000097547
|
trpc6b
|
transient receptor potential cation channel, subfamily C, member 6b |
| chr3_+_54047342 | 1.42 |
ENSDART00000178486
|
olfm2a
|
olfactomedin 2a |
| chr25_+_33063762 | 1.39 |
ENSDART00000189974
|
tln2b
|
talin 2b |
| chr9_+_18572970 | 1.37 |
ENSDART00000114860
|
lacc1
|
laccase (multicopper oxidoreductase) domain containing 1 |
| chr15_+_29088426 | 1.36 |
ENSDART00000187290
|
si:ch211-137a8.4
|
si:ch211-137a8.4 |
| chr4_-_5863906 | 1.35 |
ENSDART00000169424
|
slc5a8
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 8 |
| chr8_-_45838481 | 1.33 |
ENSDART00000148206
ENSDART00000170485 |
ogdha
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase a (lipoamide) |
| chr13_+_6092135 | 1.33 |
ENSDART00000162738
|
fam120b
|
family with sequence similarity 120B |
| chr5_-_42883761 | 1.32 |
ENSDART00000167374
|
BX323596.2
|
|
| chr18_+_17660402 | 1.32 |
ENSDART00000143475
|
cpne2
|
copine II |
| chr15_-_23485752 | 1.29 |
ENSDART00000152706
|
nlrx1
|
NLR family member X1 |
| chr19_-_12648122 | 1.29 |
ENSDART00000151184
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr9_+_219124 | 1.29 |
ENSDART00000161484
|
map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr19_+_24596324 | 1.28 |
ENSDART00000052420
|
si:dkeyp-92c9.2
|
si:dkeyp-92c9.2 |
| chr22_-_16180467 | 1.24 |
ENSDART00000171331
ENSDART00000185607 |
vcam1b
|
vascular cell adhesion molecule 1b |
| chr5_-_4245186 | 1.23 |
ENSDART00000145912
|
si:ch211-283g2.3
|
si:ch211-283g2.3 |
| chr8_-_46060685 | 1.21 |
ENSDART00000121902
|
pimr188
|
Pim proto-oncogene, serine/threonine kinase, related 188 |
| chr20_+_37294112 | 1.19 |
ENSDART00000076293
ENSDART00000140450 |
cx23
|
connexin 23 |
| chr11_-_17964525 | 1.18 |
ENSDART00000018948
|
cishb
|
cytokine inducible SH2-containing protein b |
| chr1_+_33322174 | 1.18 |
ENSDART00000140435
|
mxra5a
|
matrix-remodelling associated 5a |
| chr8_-_47212686 | 1.16 |
ENSDART00000040370
|
pimr189
|
Pim proto-oncogene, serine/threonine kinase, related 189 |
| chr14_+_22132388 | 1.15 |
ENSDART00000109065
|
ccng1
|
cyclin G1 |
| chr7_+_53254234 | 1.14 |
ENSDART00000169830
|
trip4
|
thyroid hormone receptor interactor 4 |
| chr6_+_16895685 | 1.09 |
ENSDART00000154090
|
pimr28
|
Pim proto-oncogene, serine/threonine kinase, related 28 |
| chr7_+_29009161 | 1.09 |
ENSDART00000166458
|
CR376738.1
|
|
| chr7_-_38687431 | 1.09 |
ENSDART00000074490
|
aplnr2
|
apelin receptor 2 |
| chr17_+_46864116 | 1.07 |
ENSDART00000156250
|
pimr27
|
Pim proto-oncogene, serine/threonine kinase, related 27 |
| chr7_+_29890292 | 1.06 |
ENSDART00000170403
ENSDART00000168600 |
tln2a
|
talin 2a |
| chr19_+_15443063 | 1.02 |
ENSDART00000151732
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr2_-_42109575 | 1.01 |
ENSDART00000075551
|
adhfe1
|
alcohol dehydrogenase, iron containing, 1 |
| chr22_-_9861531 | 1.01 |
ENSDART00000193197
|
si:dkey-253d23.2
|
si:dkey-253d23.2 |
| chr10_+_36299541 | 0.99 |
ENSDART00000166138
|
or106-7
|
odorant receptor, family G, subfamily 106, member 7 |
| chr24_+_13635387 | 0.99 |
ENSDART00000105997
|
trpa1b
|
transient receptor potential cation channel, subfamily A, member 1b |
| chr7_-_13362590 | 0.94 |
ENSDART00000091616
|
sdhaf2
|
succinate dehydrogenase complex assembly factor 2 |
| chr4_-_57572054 | 0.93 |
ENSDART00000158610
|
pimr168
|
Pim proto-oncogene, serine/threonine kinase, related 168 |
| chr13_-_25284716 | 0.93 |
ENSDART00000039828
|
vcla
|
vinculin a |
| chr2_-_3611960 | 0.92 |
ENSDART00000184579
|
pter
|
phosphotriesterase related |
| chr10_+_44700103 | 0.92 |
ENSDART00000165999
|
scarb1
|
scavenger receptor class B, member 1 |
| chr13_-_9335891 | 0.92 |
ENSDART00000080637
|
BX901922.1
|
|
| chr25_+_13620555 | 0.91 |
ENSDART00000163642
|
si:ch211-172l8.4
|
si:ch211-172l8.4 |
| chr21_+_37090585 | 0.91 |
ENSDART00000182971
|
znf346
|
zinc finger protein 346 |
| chr15_-_41290415 | 0.90 |
ENSDART00000152157
|
si:dkey-75b17.1
|
si:dkey-75b17.1 |
| chr15_-_25039083 | 0.88 |
ENSDART00000154409
|
pimr190
|
Pim proto-oncogene, serine/threonine kinase, related 190 |
| chr8_+_10920205 | 0.86 |
ENSDART00000179742
|
BX321886.1
|
|
| chr15_-_31401361 | 0.84 |
ENSDART00000124360
|
or111-7
|
odorant receptor, family D, subfamily 111, member 7 |
| chr17_+_12255642 | 0.83 |
ENSDART00000155175
|
pimr175
|
Pim proto-oncogene, serine/threonine kinase, related 175 |
| chr4_+_34599803 | 0.83 |
ENSDART00000134088
|
pimr168
|
Pim proto-oncogene, serine/threonine kinase, related 168 |
| chr20_-_54508650 | 0.82 |
ENSDART00000147642
|
CABZ01118770.1
|
|
| chr12_+_46742805 | 0.80 |
ENSDART00000192236
|
plaub
|
plasminogen activator, urokinase b |
| chr7_+_17560808 | 0.76 |
ENSDART00000159699
ENSDART00000165724 ENSDART00000129017 ENSDART00000101955 |
nitr1b
|
novel immune-type receptor 1b |
| chr7_+_17303944 | 0.76 |
ENSDART00000163505
ENSDART00000162169 ENSDART00000161291 |
nitr9
|
novel immune-type receptor 9 |
| chr4_-_57571688 | 0.76 |
ENSDART00000160179
|
pimr168
|
Pim proto-oncogene, serine/threonine kinase, related 168 |
| chr17_-_43517542 | 0.74 |
ENSDART00000133665
|
mrpl35
|
mitochondrial ribosomal protein L35 |
| chr16_+_25126935 | 0.73 |
ENSDART00000058945
|
zgc:92590
|
zgc:92590 |
| chr7_+_33133547 | 0.72 |
ENSDART00000191967
|
si:ch211-194p6.7
|
si:ch211-194p6.7 |
| chr16_-_52879741 | 0.69 |
ENSDART00000166470
|
TPPP
|
tubulin polymerization promoting protein |
| chr12_+_22404108 | 0.65 |
ENSDART00000153055
|
hdlbpb
|
high density lipoprotein binding protein b |
| chr6_-_16804001 | 0.63 |
ENSDART00000155398
|
pimr40
|
Pim proto-oncogene, serine/threonine kinase, related 40 |
| chr17_+_6536152 | 0.62 |
ENSDART00000062952
ENSDART00000121789 |
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr16_+_34522977 | 0.62 |
ENSDART00000144069
|
paqr7b
|
progestin and adipoQ receptor family member VII, b |
| chr23_-_37432459 | 0.62 |
ENSDART00000188916
|
zmp:0000001088
|
zmp:0000001088 |
| chr22_-_10774735 | 0.59 |
ENSDART00000081156
|
timm13
|
translocase of inner mitochondrial membrane 13 homolog (yeast) |
| chr17_-_46817295 | 0.55 |
ENSDART00000155904
|
pimr24
|
Pim proto-oncogene, serine/threonine kinase, related 24 |
| chr15_-_29556757 | 0.55 |
ENSDART00000060049
|
hspa13
|
heat shock protein 70 family, member 13 |
| chr6_-_17266221 | 0.54 |
ENSDART00000155448
|
pimr18
|
Pim proto-oncogene, serine/threonine kinase, related 18 |
| chr6_-_17052116 | 0.51 |
ENSDART00000156825
|
pimr45
|
Pim proto-oncogene, serine/threonine kinase, related 45 |
| chr2_+_44426609 | 0.48 |
ENSDART00000112711
ENSDART00000154188 |
kcnab1b
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 b |
| chr17_-_46933567 | 0.48 |
ENSDART00000157274
|
pimr25
|
Pim proto-oncogene, serine/threonine kinase, related 25 |
| chr1_-_10647484 | 0.48 |
ENSDART00000164541
ENSDART00000188958 ENSDART00000190904 |
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr6_-_16735402 | 0.48 |
ENSDART00000154216
|
pimr44
|
Pim proto-oncogene, serine/threonine kinase, related 44 |
| chr5_-_42071505 | 0.47 |
ENSDART00000137224
ENSDART00000193721 |
cxcl11.7
cenpv
|
chemokine (C-X-C motif) ligand 11, duplicate 7 centromere protein V |
| chr13_+_2861265 | 0.47 |
ENSDART00000170602
ENSDART00000171687 |
XPO5
|
si:ch211-233m11.2 |
| chr15_-_25010400 | 0.46 |
ENSDART00000155236
|
si:ch211-198e20.10
|
si:ch211-198e20.10 |
| chr24_+_13635108 | 0.44 |
ENSDART00000183008
|
trpa1b
|
transient receptor potential cation channel, subfamily A, member 1b |
| chr1_-_493218 | 0.43 |
ENSDART00000031635
|
ercc5
|
excision repair cross-complementation group 5 |
| chr10_+_44699734 | 0.41 |
ENSDART00000167952
ENSDART00000158681 ENSDART00000190188 ENSDART00000168276 |
scarb1
|
scavenger receptor class B, member 1 |
| chr17_-_6536466 | 0.36 |
ENSDART00000188735
|
cenpo
|
centromere protein O |
| chr25_-_30394746 | 0.35 |
ENSDART00000185346
|
si:ch211-93f2.1
|
si:ch211-93f2.1 |
| chr1_-_23588996 | 0.35 |
ENSDART00000102600
|
lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr13_-_17464654 | 0.34 |
ENSDART00000140312
|
lrmda
|
leucine rich melanocyte differentiation associated |
| chr4_-_18595525 | 0.34 |
ENSDART00000049061
|
cdkn1ba
|
cyclin-dependent kinase inhibitor 1Ba |
| chr20_+_28803977 | 0.33 |
ENSDART00000153351
ENSDART00000038149 |
fntb
|
farnesyltransferase, CAAX box, beta |
| chr12_+_48480632 | 0.32 |
ENSDART00000158157
|
arhgap44
|
Rho GTPase activating protein 44 |
| chr17_-_6536305 | 0.31 |
ENSDART00000154855
|
cenpo
|
centromere protein O |
| chr1_-_10647307 | 0.29 |
ENSDART00000103548
|
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr3_-_32541033 | 0.29 |
ENSDART00000151476
ENSDART00000055324 |
rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr6_+_41503854 | 0.29 |
ENSDART00000136538
ENSDART00000140108 ENSDART00000084861 |
cish
|
cytokine inducible SH2-containing protein |
| chr3_-_26904774 | 0.29 |
ENSDART00000103690
|
clec16a
|
C-type lectin domain containing 16A |
| chr8_+_25959940 | 0.28 |
ENSDART00000143011
ENSDART00000140626 |
si:dkey-72l14.4
|
si:dkey-72l14.4 |
| chr17_-_24439672 | 0.27 |
ENSDART00000155020
|
wdpcp
|
WD repeat containing planar cell polarity effector |
| chr23_-_1571682 | 0.26 |
ENSDART00000013635
|
fbxo30b
|
F-box protein 30b |
| chr11_+_36409457 | 0.26 |
ENSDART00000077641
|
cyb561d1
|
cytochrome b561 family, member D1 |
| chr3_-_6608342 | 0.25 |
ENSDART00000161188
|
si:ch73-59f11.3
|
si:ch73-59f11.3 |
| chr7_+_65813301 | 0.24 |
ENSDART00000187449
ENSDART00000179834 ENSDART00000082740 ENSDART00000178627 |
tead1b
|
TEA domain family member 1b |
| chr12_-_23365737 | 0.24 |
ENSDART00000170376
|
mpp7a
|
membrane protein, palmitoylated 7a (MAGUK p55 subfamily member 7) |
| chr15_-_6966221 | 0.23 |
ENSDART00000165487
ENSDART00000027657 |
mrps22
|
mitochondrial ribosomal protein S22 |
| chr8_+_247163 | 0.23 |
ENSDART00000122378
|
cep120
|
centrosomal protein 120 |
| chr20_+_52458765 | 0.22 |
ENSDART00000057980
|
tsta3
|
tissue specific transplantation antigen P35B |
| chr20_+_28803642 | 0.22 |
ENSDART00000188526
|
fntb
|
farnesyltransferase, CAAX box, beta |
| chr21_-_27010796 | 0.21 |
ENSDART00000065398
ENSDART00000144342 ENSDART00000126542 |
ppp1r14ba
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Ba |
| chr4_+_13586455 | 0.21 |
ENSDART00000187230
|
tnpo3
|
transportin 3 |
| chr4_-_49952636 | 0.18 |
ENSDART00000157941
|
si:dkey-156k2.3
|
si:dkey-156k2.3 |
| chr14_-_6987649 | 0.16 |
ENSDART00000060990
|
eif4ebp3l
|
eukaryotic translation initiation factor 4E binding protein 3, like |
| chr18_-_20444296 | 0.15 |
ENSDART00000132993
|
kif23
|
kinesin family member 23 |
| chr1_+_57040472 | 0.15 |
ENSDART00000181365
|
si:ch211-1f22.16
|
si:ch211-1f22.16 |
| chr11_+_13024002 | 0.13 |
ENSDART00000104113
|
btf3l4
|
basic transcription factor 3-like 4 |
| chr15_+_2857556 | 0.10 |
ENSDART00000157758
|
mre11a
|
MRE11 homolog A, double strand break repair nuclease |
| chr24_-_18477672 | 0.08 |
ENSDART00000126928
ENSDART00000158393 |
si:dkey-73n8.3
|
si:dkey-73n8.3 |
| chr15_-_25083200 | 0.07 |
ENSDART00000156053
|
pimr191
|
Pim proto-oncogene, serine/threonine kinase, related 191 |
| chr21_+_22400951 | 0.07 |
ENSDART00000134592
|
lmbrd2b
|
LMBR1 domain containing 2b |
| chr11_+_30314885 | 0.06 |
ENSDART00000187418
ENSDART00000123244 |
ugt1b2
|
UDP glucuronosyltransferase 1 family, polypeptide B2 |
| chr4_-_685412 | 0.04 |
ENSDART00000168167
|
rtcb
|
RNA 2',3'-cyclic phosphate and 5'-OH ligase |
| chr17_+_11372531 | 0.04 |
ENSDART00000130975
ENSDART00000149366 |
timm9
|
translocase of inner mitochondrial membrane 9 homolog |
Network of associatons between targets according to the STRING database.
First level regulatory network of stat5a+stat5b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0042245 | RNA repair(GO:0042245) |
| 0.6 | 2.5 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.5 | 1.6 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.5 | 2.5 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.5 | 1.5 | GO:0021577 | hindbrain structural organization(GO:0021577) dorsal fin morphogenesis(GO:0035142) cell motility involved in somitogenic axis elongation(GO:0090247) cell migration involved in somitogenic axis elongation(GO:0090248) |
| 0.5 | 1.4 | GO:0050968 | chemosensory behavior(GO:0007635) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.4 | 1.3 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.3 | 3.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.2 | 2.0 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.2 | 2.3 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.2 | 3.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 0.9 | GO:0097032 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.2 | 2.4 | GO:0050482 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.2 | 1.8 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.2 | 0.6 | GO:0051182 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.2 | 0.9 | GO:0086013 | membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.1 | 0.4 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 | 0.5 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 3.2 | GO:0006595 | polyamine metabolic process(GO:0006595) |
| 0.1 | 0.5 | GO:1901380 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 | 0.6 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 2.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 1.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 1.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 1.5 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 1.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 1.8 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 0.8 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.1 | 0.3 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 0.2 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.1 | 1.3 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 1.5 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.3 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 1.6 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 1.3 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.0 | 2.1 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 1.3 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 1.5 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.2 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.0 | 1.7 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 3.8 | GO:0006956 | complement activation(GO:0006956) |
| 0.0 | 13.3 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.5 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.6 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 1.1 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.2 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 1.6 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.3 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.8 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 0.3 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 3.0 | GO:0040008 | regulation of growth(GO:0040008) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 3.5 | GO:0050767 | regulation of neurogenesis(GO:0050767) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.5 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.3 | 2.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 1.7 | GO:0071203 | WASH complex(GO:0071203) |
| 0.3 | 1.3 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.2 | 0.6 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 0.7 | GO:0031511 | Mis6-Sim4 complex(GO:0031511) |
| 0.1 | 0.5 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 1.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 1.5 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 0.3 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.1 | 1.5 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 2.5 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 1.3 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 3.8 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.6 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 2.4 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.9 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 1.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 3.8 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 14.3 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 1.5 | GO:0034703 | cation channel complex(GO:0034703) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.8 | GO:1990931 | oxidative DNA demethylase activity(GO:0035516) DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.8 | 3.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.5 | 2.5 | GO:0015140 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.4 | 1.7 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.3 | 1.3 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.3 | 1.5 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.3 | 2.3 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.3 | 3.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.2 | 2.5 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.2 | 1.1 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.2 | 1.8 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.2 | 1.0 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.2 | 3.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 1.3 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.2 | 0.6 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.1 | 1.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 1.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.5 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 1.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 2.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 2.7 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 2.3 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.1 | 1.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 1.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 1.4 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 1.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 2.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.3 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.0 | 0.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 1.5 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 1.6 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.7 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 1.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.5 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.5 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 1.4 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 1.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.9 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 14.3 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.6 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 1.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 3.6 | GO:0015631 | tubulin binding(GO:0015631) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 0.3 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.1 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 2.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 3.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 1.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.5 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 1.0 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.4 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.3 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |