Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
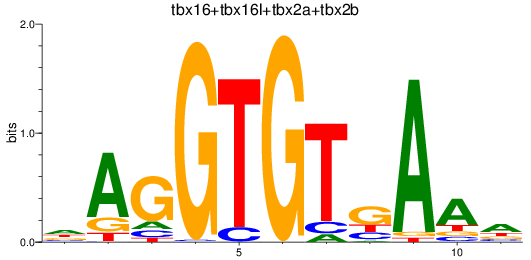
Results for tbr1b_tbx16+tbx16l+tbx2a+tbx2b
Z-value: 1.14
Transcription factors associated with tbr1b_tbx16+tbx16l+tbx2a+tbx2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tbr1b
|
ENSDARG00000004712 | T-box brain transcription factor 1b |
|
tbx2b
|
ENSDARG00000006120 | T-box transcription factor 2b |
|
tbx16l
|
ENSDARG00000006939 | T-box transcription factor 16, like |
|
tbx16
|
ENSDARG00000007329 | T-box transcription factor 16 |
|
tbx2a
|
ENSDARG00000018025 | T-box transcription factor 2a |
|
tbx2a
|
ENSDARG00000109541 | T-box transcription factor 2a |
|
tbx2b
|
ENSDARG00000116135 | T-box transcription factor 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tbx16 | dr11_v1_chr8_-_51753604_51753604 | -0.77 | 1.6e-19 | Click! |
| tbr1b | dr11_v1_chr9_-_51436377_51436377 | -0.69 | 2.0e-14 | Click! |
| tbx6l | dr11_v1_chr5_+_42280372_42280372 | -0.25 | 1.4e-02 | Click! |
| tbx2a | dr11_v1_chr5_-_56513825_56513825 | 0.15 | 1.5e-01 | Click! |
| tbx2b | dr11_v1_chr15_+_27364394_27364394 | 0.08 | 4.2e-01 | Click! |
Activity profile of tbr1b_tbx16+tbx16l+tbx2a+tbx2b motif
Sorted Z-values of tbr1b_tbx16+tbx16l+tbx2a+tbx2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_51846224 | 28.15 |
ENSDART00000184663
|
LT631684.2
|
|
| chr7_+_7048245 | 15.40 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr7_+_6969909 | 12.59 |
ENSDART00000189886
|
actn3b
|
actinin alpha 3b |
| chr17_+_26965351 | 11.42 |
ENSDART00000114215
ENSDART00000147192 |
grhl3
|
grainyhead-like transcription factor 3 |
| chr7_+_69841017 | 11.22 |
ENSDART00000169107
|
FO818704.1
|
|
| chr9_-_42873700 | 10.73 |
ENSDART00000125953
|
ttn.1
|
titin, tandem duplicate 1 |
| chr3_+_32526263 | 10.62 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr19_+_791538 | 9.72 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr23_+_23232136 | 9.11 |
ENSDART00000126479
ENSDART00000187764 |
plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr13_-_15702672 | 8.63 |
ENSDART00000144445
ENSDART00000168950 |
ckba
|
creatine kinase, brain a |
| chr14_+_21106444 | 8.61 |
ENSDART00000075744
ENSDART00000132363 |
aldob
|
aldolase b, fructose-bisphosphate |
| chr11_-_25257045 | 8.58 |
ENSDART00000130477
|
snai1a
|
snail family zinc finger 1a |
| chr6_+_56141852 | 7.95 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr23_-_20100436 | 7.93 |
ENSDART00000143445
|
si:dkey-32e6.6
|
si:dkey-32e6.6 |
| chr3_-_61181018 | 7.90 |
ENSDART00000187970
|
pvalb4
|
parvalbumin 4 |
| chr21_-_22928214 | 7.83 |
ENSDART00000182760
|
dub
|
duboraya |
| chr3_-_32603191 | 7.79 |
ENSDART00000150997
|
si:ch73-248e21.7
|
si:ch73-248e21.7 |
| chr7_+_39446247 | 7.73 |
ENSDART00000033610
ENSDART00000099015 |
tnnt3b
|
troponin T type 3b (skeletal, fast) |
| chr15_+_32711663 | 7.73 |
ENSDART00000157854
ENSDART00000167515 |
postnb
|
periostin, osteoblast specific factor b |
| chr18_-_7539166 | 7.62 |
ENSDART00000133541
|
si:dkey-30c15.2
|
si:dkey-30c15.2 |
| chr2_+_36007449 | 7.51 |
ENSDART00000161837
|
lamc2
|
laminin, gamma 2 |
| chr22_+_661711 | 7.36 |
ENSDART00000113795
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr11_-_25257595 | 7.35 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr12_+_6002715 | 7.31 |
ENSDART00000114961
|
si:ch211-131k2.3
|
si:ch211-131k2.3 |
| chr1_-_49521407 | 7.28 |
ENSDART00000189845
ENSDART00000143474 |
zp3c
|
zona pellucida glycoprotein 3c |
| chr18_+_29156827 | 7.23 |
ENSDART00000137587
ENSDART00000135633 |
ppfibp2a
|
PTPRF interacting protein, binding protein 2a (liprin beta 2) |
| chr22_+_26600834 | 7.10 |
ENSDART00000157411
|
adcy9
|
adenylate cyclase 9 |
| chr5_+_43006422 | 7.03 |
ENSDART00000009182
|
aqp3a
|
aquaporin 3a |
| chr7_+_38750871 | 6.98 |
ENSDART00000114238
ENSDART00000052325 ENSDART00000137001 |
f2
|
coagulation factor II (thrombin) |
| chr7_-_52417777 | 6.93 |
ENSDART00000110265
|
myzap
|
myocardial zonula adherens protein |
| chr25_-_27819838 | 6.84 |
ENSDART00000067106
|
lmod2a
|
leiomodin 2 (cardiac) a |
| chr13_+_33606739 | 6.72 |
ENSDART00000026464
|
cfl1l
|
cofilin 1 (non-muscle), like |
| chr9_-_443451 | 6.70 |
ENSDART00000165642
|
si:dkey-11f4.14
|
si:dkey-11f4.14 |
| chr19_+_30990815 | 6.66 |
ENSDART00000134645
|
sync
|
syncoilin, intermediate filament protein |
| chr19_-_7420867 | 6.58 |
ENSDART00000081741
|
rab25a
|
RAB25, member RAS oncogene family a |
| chr19_+_7735157 | 6.42 |
ENSDART00000186717
|
tuft1b
|
tuftelin 1b |
| chr21_-_11575626 | 6.15 |
ENSDART00000133171
|
cast
|
calpastatin |
| chr7_+_3357973 | 6.13 |
ENSDART00000172853
|
si:ch211-285c6.5
|
si:ch211-285c6.5 |
| chr10_-_1961930 | 6.08 |
ENSDART00000122446
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr12_-_10567188 | 5.87 |
ENSDART00000144283
|
myof
|
myoferlin |
| chr24_+_35975398 | 5.72 |
ENSDART00000173058
|
obscnb
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF b |
| chr15_+_32711172 | 5.63 |
ENSDART00000163936
ENSDART00000168135 |
postnb
|
periostin, osteoblast specific factor b |
| chr7_+_65240227 | 5.61 |
ENSDART00000168287
|
bco1l
|
beta-carotene oxygenase 1, like |
| chr14_+_17376940 | 5.53 |
ENSDART00000054590
ENSDART00000010148 |
spon2b
|
spondin 2b, extracellular matrix protein |
| chr14_+_48862987 | 5.51 |
ENSDART00000167810
|
zgc:154054
|
zgc:154054 |
| chr20_-_26846028 | 5.48 |
ENSDART00000136687
|
mylk4b
|
myosin light chain kinase family, member 4b |
| chr10_+_43994471 | 5.42 |
ENSDART00000138242
ENSDART00000186359 |
cldn5b
|
claudin 5b |
| chr18_+_7591381 | 5.42 |
ENSDART00000136313
|
si:dkeyp-1h4.6
|
si:dkeyp-1h4.6 |
| chr22_+_25693295 | 5.39 |
ENSDART00000123888
ENSDART00000150783 |
si:dkeyp-98a7.4
si:dkeyp-98a7.3
|
si:dkeyp-98a7.4 si:dkeyp-98a7.3 |
| chr5_-_25037454 | 5.38 |
ENSDART00000027237
|
abo
|
ABO blood group (transferase A, alpha 1-3-N-acetylgalactosaminyltransferase; transferase B, alpha 1-3-galactosyltransferase) |
| chr23_+_9867483 | 5.33 |
ENSDART00000023099
|
slc16a7
|
solute carrier family 16, member 7 (monocarboxylic acid transporter 2) |
| chr22_+_25704430 | 5.32 |
ENSDART00000143776
|
si:dkeyp-98a7.3
|
si:dkeyp-98a7.3 |
| chr22_-_24757785 | 5.30 |
ENSDART00000078225
|
vtg5
|
vitellogenin 5 |
| chr8_+_39663612 | 5.29 |
ENSDART00000188074
|
BX005328.1
|
|
| chr2_-_30182353 | 5.25 |
ENSDART00000019149
|
rpl7
|
ribosomal protein L7 |
| chr22_+_25681911 | 5.23 |
ENSDART00000113381
|
si:dkeyp-98a7.3
|
si:dkeyp-98a7.3 |
| chr19_+_2590182 | 5.22 |
ENSDART00000162293
|
si:ch73-345f18.3
|
si:ch73-345f18.3 |
| chr7_-_19614916 | 5.15 |
ENSDART00000169029
|
zgc:194655
|
zgc:194655 |
| chr22_+_25715925 | 5.13 |
ENSDART00000150650
|
si:dkeyp-98a7.7
|
si:dkeyp-98a7.7 |
| chr17_-_8907668 | 4.97 |
ENSDART00000155996
ENSDART00000155624 |
nkl.1
|
NK-lysin tandem duplicate 1 |
| chr8_-_24252933 | 4.97 |
ENSDART00000057624
|
zgc:110353
|
zgc:110353 |
| chr22_+_25687525 | 4.95 |
ENSDART00000135717
|
si:dkeyp-98a7.3
|
si:dkeyp-98a7.3 |
| chr13_+_31757331 | 4.94 |
ENSDART00000044282
|
hif1aa
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) a |
| chr7_-_53117131 | 4.94 |
ENSDART00000169211
ENSDART00000168890 ENSDART00000172179 ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr25_+_4960509 | 4.93 |
ENSDART00000155984
|
si:ch73-265h17.4
|
si:ch73-265h17.4 |
| chr11_+_6650966 | 4.92 |
ENSDART00000131236
|
si:dkey-246j7.1
|
si:dkey-246j7.1 |
| chr10_+_3127809 | 4.92 |
ENSDART00000185500
|
ckmt2a
|
creatine kinase, mitochondrial 2a (sarcomeric) |
| chr13_+_18533005 | 4.85 |
ENSDART00000136024
|
ftr14l
|
finTRIM family, member 14-like |
| chr19_+_9305964 | 4.83 |
ENSDART00000136241
|
si:ch73-15n24.1
|
si:ch73-15n24.1 |
| chr21_+_10866421 | 4.81 |
ENSDART00000137858
|
alpk2
|
alpha-kinase 2 |
| chr13_-_9442942 | 4.78 |
ENSDART00000138833
|
grxcr1
|
glutaredoxin, cysteine rich 1 |
| chr8_-_11229523 | 4.76 |
ENSDART00000002164
|
unc45b
|
unc-45 myosin chaperone B |
| chr3_+_43086548 | 4.72 |
ENSDART00000163579
|
si:dkey-43p13.5
|
si:dkey-43p13.5 |
| chr5_+_27525477 | 4.70 |
ENSDART00000051491
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr24_-_27419198 | 4.63 |
ENSDART00000141124
|
ccl34b.4
|
chemokine (C-C motif) ligand 34b, duplicate 4 |
| chr6_-_46941245 | 4.60 |
ENSDART00000037875
|
igfn1.1
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 1 |
| chr24_-_34680956 | 4.58 |
ENSDART00000171009
|
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr11_+_37178271 | 4.56 |
ENSDART00000161771
|
itih3b
|
inter-alpha-trypsin inhibitor heavy chain 3b |
| chr12_+_10062953 | 4.47 |
ENSDART00000148689
|
slc4a1b
|
solute carrier family 4 (anion exchanger), member 1b (Diego blood group) |
| chr22_+_25710148 | 4.42 |
ENSDART00000150446
|
si:dkeyp-98a7.4
|
si:dkeyp-98a7.4 |
| chr10_-_32524035 | 4.40 |
ENSDART00000137608
|
dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr2_-_42234484 | 4.40 |
ENSDART00000132617
ENSDART00000136690 ENSDART00000141358 |
apom
|
apolipoprotein M |
| chr23_-_29003864 | 4.38 |
ENSDART00000148257
|
casz1
|
castor zinc finger 1 |
| chr9_-_40011673 | 4.37 |
ENSDART00000184726
|
IKZF2
|
si:zfos-1425h8.1 |
| chr17_+_51517750 | 4.37 |
ENSDART00000180896
ENSDART00000193528 |
pxdn
|
peroxidasin |
| chr4_-_11580948 | 4.35 |
ENSDART00000049066
|
net1
|
neuroepithelial cell transforming 1 |
| chr16_-_32233463 | 4.35 |
ENSDART00000102016
|
calhm6
|
calcium homeostasis modulator family member 6 |
| chr7_-_58776400 | 4.33 |
ENSDART00000167433
|
sox17
|
SRY (sex determining region Y)-box 17 |
| chr5_-_4204580 | 4.32 |
ENSDART00000049197
ENSDART00000132130 |
si:ch211-283g2.1
|
si:ch211-283g2.1 |
| chr5_-_44843738 | 4.31 |
ENSDART00000003926
|
fbp1a
|
fructose-1,6-bisphosphatase 1a |
| chr25_+_36292057 | 4.30 |
ENSDART00000152329
|
bmb
|
brambleberry |
| chr19_+_917852 | 4.27 |
ENSDART00000082466
|
tgfbr2a
|
transforming growth factor beta receptor 2a |
| chr22_+_25720725 | 4.24 |
ENSDART00000150778
|
si:dkeyp-98a7.8
|
si:dkeyp-98a7.8 |
| chr21_-_34951265 | 4.24 |
ENSDART00000135222
|
lipia
|
lipase, member Ia |
| chr15_-_5580093 | 4.24 |
ENSDART00000143726
|
wdr62
|
WD repeat domain 62 |
| chr22_+_1123110 | 4.22 |
ENSDART00000171017
|
si:ch1073-181h11.2
|
si:ch1073-181h11.2 |
| chr1_-_6028876 | 4.22 |
ENSDART00000168117
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr24_-_36593876 | 4.18 |
ENSDART00000160901
|
CABZ01055365.1
|
|
| chr18_-_48550426 | 4.17 |
ENSDART00000145189
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr9_-_21970067 | 4.16 |
ENSDART00000009920
|
lmo7a
|
LIM domain 7a |
| chr5_-_38170996 | 4.10 |
ENSDART00000145805
|
si:ch211-284e13.12
|
si:ch211-284e13.12 |
| chr13_-_51922290 | 4.04 |
ENSDART00000168648
|
srfb
|
serum response factor b |
| chr13_-_41908583 | 4.03 |
ENSDART00000136515
|
ipmka
|
inositol polyphosphate multikinase a |
| chr7_+_65673885 | 3.99 |
ENSDART00000169182
|
parvab
|
parvin, alpha b |
| chr1_+_8521323 | 3.98 |
ENSDART00000121439
ENSDART00000103626 ENSDART00000141283 |
mief2
|
mitochondrial elongation factor 2 |
| chr16_+_13993285 | 3.98 |
ENSDART00000139130
ENSDART00000130353 |
si:dkey-85k15.7
fdps
|
si:dkey-85k15.7 farnesyl diphosphate synthase (farnesyl pyrophosphate synthetase, dimethylallyltranstransferase, geranyltranstransferase) |
| chr21_+_5531138 | 3.97 |
ENSDART00000163825
|
ly6m6
|
lymphocyte antigen 6 family member M6 |
| chr2_+_45081489 | 3.96 |
ENSDART00000123966
|
chrng
|
cholinergic receptor, nicotinic, gamma |
| chr21_+_28445052 | 3.95 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr12_-_14143344 | 3.94 |
ENSDART00000152742
|
buc2l
|
bucky ball 2-like |
| chr13_+_1799003 | 3.93 |
ENSDART00000029343
|
col21a1
|
collagen, type XXI, alpha 1 |
| chr5_-_32505109 | 3.91 |
ENSDART00000188219
|
ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr10_+_33171501 | 3.90 |
ENSDART00000159666
|
myl10
|
myosin, light chain 10, regulatory |
| chr16_+_29492937 | 3.90 |
ENSDART00000011497
|
ctsk
|
cathepsin K |
| chr16_-_12723738 | 3.89 |
ENSDART00000080414
|
sbk3
|
SH3 domain binding kinase family, member 3 |
| chr7_+_2797795 | 3.86 |
ENSDART00000161208
|
si:ch211-217i17.1
|
si:ch211-217i17.1 |
| chr1_+_51615672 | 3.85 |
ENSDART00000165117
|
zgc:165656
|
zgc:165656 |
| chr5_-_42878178 | 3.85 |
ENSDART00000162981
|
CXCL11 (1 of many)
|
C-X-C motif chemokine ligand 11 |
| chr18_+_8340886 | 3.85 |
ENSDART00000081132
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr8_+_36142734 | 3.84 |
ENSDART00000159361
ENSDART00000161194 |
mhc2b
|
major histocompatibility complex class II integral membrane beta chain gene |
| chr24_-_18919562 | 3.83 |
ENSDART00000144244
ENSDART00000106188 ENSDART00000182518 |
cpa6
|
carboxypeptidase A6 |
| chr14_+_97017 | 3.83 |
ENSDART00000159300
ENSDART00000169523 |
mcm7
|
minichromosome maintenance complex component 7 |
| chr6_+_36877968 | 3.81 |
ENSDART00000155187
|
traf3ip2l
|
TRAF3 interacting protein 2-like |
| chr22_-_35330532 | 3.78 |
ENSDART00000172654
|
FO818743.1
|
|
| chr10_-_25816558 | 3.77 |
ENSDART00000017240
|
postna
|
periostin, osteoblast specific factor a |
| chr9_+_8851819 | 3.69 |
ENSDART00000160004
|
col4a2
|
collagen, type IV, alpha 2 |
| chr17_+_34186632 | 3.68 |
ENSDART00000014306
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr24_-_26369185 | 3.65 |
ENSDART00000080039
|
lrrc31
|
leucine rich repeat containing 31 |
| chr7_+_13988075 | 3.65 |
ENSDART00000186812
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr15_-_43164591 | 3.64 |
ENSDART00000171305
|
ap1s3a
|
adaptor-related protein complex 1, sigma 3 subunit, a |
| chr12_+_13244149 | 3.62 |
ENSDART00000186984
ENSDART00000105896 |
atp2a1l
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1, like |
| chr19_-_34999379 | 3.60 |
ENSDART00000051751
|
zgc:113424
|
zgc:113424 |
| chr6_+_36839509 | 3.60 |
ENSDART00000190605
ENSDART00000104160 |
zgc:110788
|
zgc:110788 |
| chr22_+_25734180 | 3.59 |
ENSDART00000143367
|
si:dkeyp-98a7.9
|
si:dkeyp-98a7.9 |
| chr13_-_25767210 | 3.58 |
ENSDART00000131792
|
pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr17_-_5583345 | 3.57 |
ENSDART00000035944
|
clic5a
|
chloride intracellular channel 5a |
| chr15_-_47848544 | 3.57 |
ENSDART00000098711
|
eif3k
|
eukaryotic translation initiation factor 3, subunit K |
| chr18_+_54354 | 3.50 |
ENSDART00000097163
|
zgc:158482
|
zgc:158482 |
| chr7_-_5070794 | 3.48 |
ENSDART00000097877
|
ltb4r2a
|
leukotriene B4 receptor 2a |
| chr19_-_5332784 | 3.48 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr22_-_9649627 | 3.46 |
ENSDART00000164721
|
si:dkey-286j17.4
|
si:dkey-286j17.4 |
| chr3_-_23643751 | 3.46 |
ENSDART00000078425
ENSDART00000140264 |
eve1
|
even-skipped-like1 |
| chr5_-_68022631 | 3.44 |
ENSDART00000143199
|
wasf3a
|
WAS protein family, member 3a |
| chr22_-_17652112 | 3.42 |
ENSDART00000189205
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr16_-_20312146 | 3.41 |
ENSDART00000134980
|
si:dkeyp-86h10.3
|
si:dkeyp-86h10.3 |
| chr16_-_54455573 | 3.41 |
ENSDART00000075275
|
pklr
|
pyruvate kinase L/R |
| chr21_-_3700334 | 3.41 |
ENSDART00000137844
|
atp8b1
|
ATPase phospholipid transporting 8B1 |
| chr3_-_39171968 | 3.39 |
ENSDART00000154494
|
si:dkeyp-57f11.2
|
si:dkeyp-57f11.2 |
| chr8_-_25327809 | 3.39 |
ENSDART00000137242
|
eps8l3b
|
EPS8-like 3b |
| chr10_+_38610741 | 3.38 |
ENSDART00000126444
|
mmp13a
|
matrix metallopeptidase 13a |
| chr17_+_38255105 | 3.38 |
ENSDART00000005296
|
nkx2.9
|
NK2 transcription factor related, locus 9 (Drosophila) |
| chr10_-_13116337 | 3.37 |
ENSDART00000164568
|
musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr15_-_1590858 | 3.37 |
ENSDART00000081875
|
nnr
|
nanor |
| chr11_+_27274355 | 3.36 |
ENSDART00000113707
|
fbln2
|
fibulin 2 |
| chr3_+_32526799 | 3.36 |
ENSDART00000185755
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr16_+_38201840 | 3.35 |
ENSDART00000044971
|
myo1eb
|
myosin IE, b |
| chr13_-_27354003 | 3.33 |
ENSDART00000101479
ENSDART00000044652 |
ddx43
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 43 |
| chr19_-_20270178 | 3.32 |
ENSDART00000144891
ENSDART00000090883 |
gpnmb
|
glycoprotein (transmembrane) nmb |
| chr2_+_38161318 | 3.31 |
ENSDART00000044264
|
mmp14b
|
matrix metallopeptidase 14b (membrane-inserted) |
| chr7_-_38638809 | 3.31 |
ENSDART00000144341
|
c6ast4
|
six-cysteine containing astacin protease 4 |
| chr10_+_38417512 | 3.31 |
ENSDART00000112457
|
samsn1b
|
SAM domain, SH3 domain and nuclear localisation signals 1b |
| chr4_+_14981854 | 3.30 |
ENSDART00000067046
|
cax1
|
cation/H+ exchanger protein 1 |
| chr20_+_15600167 | 3.25 |
ENSDART00000171991
|
faslg
|
Fas ligand (TNF superfamily, member 6) |
| chr2_-_58257624 | 3.25 |
ENSDART00000098940
|
foxl2b
|
forkhead box L2b |
| chr6_+_41039166 | 3.23 |
ENSDART00000125659
|
entpd8
|
ectonucleoside triphosphate diphosphohydrolase 8 |
| chr10_-_11376491 | 3.22 |
ENSDART00000145346
|
plac8.2
|
placenta-specific 8, tandem duplicate 2 |
| chr16_+_21790870 | 3.21 |
ENSDART00000155039
|
trim108
|
tripartite motif containing 108 |
| chr6_-_46861676 | 3.21 |
ENSDART00000188712
ENSDART00000190148 |
igfn1.3
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 3 |
| chr20_-_33961697 | 3.19 |
ENSDART00000061765
|
selp
|
selectin P |
| chr15_+_7054754 | 3.16 |
ENSDART00000149800
|
foxl2a
|
forkhead box L2a |
| chr11_-_7380674 | 3.16 |
ENSDART00000014979
ENSDART00000103418 |
vtg3
|
vitellogenin 3, phosvitinless |
| chr5_+_33498253 | 3.16 |
ENSDART00000140993
|
ms4a17c.2
|
membrane-spanning 4-domains, subfamily A, member 17c.2 |
| chr2_+_2470687 | 3.15 |
ENSDART00000184024
ENSDART00000061955 |
myl13
|
myosin, light chain 13 |
| chr10_+_252425 | 3.14 |
ENSDART00000059478
|
lrrc32
|
leucine rich repeat containing 32 |
| chr21_+_21374277 | 3.14 |
ENSDART00000079431
|
rtn2b
|
reticulon 2b |
| chr10_-_1961576 | 3.13 |
ENSDART00000042441
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr16_+_29516098 | 3.12 |
ENSDART00000174895
|
ctss2.2
|
cathepsin S, ortholog 2, tandem duplicate 2 |
| chr5_-_69180587 | 3.09 |
ENSDART00000156681
ENSDART00000160753 |
zgc:171967
|
zgc:171967 |
| chr20_-_51946052 | 3.09 |
ENSDART00000074325
|
dusp10
|
dual specificity phosphatase 10 |
| chr17_-_8312923 | 3.09 |
ENSDART00000064678
|
lft2
|
lefty2 |
| chr16_+_23978978 | 3.08 |
ENSDART00000058964
ENSDART00000135084 |
apoa2
|
apolipoprotein A-II |
| chr8_+_50531709 | 3.07 |
ENSDART00000193352
|
PEBP4
|
phosphatidylethanolamine binding protein 4 |
| chr18_+_27300700 | 3.07 |
ENSDART00000140444
|
plekha7a
|
pleckstrin homology domain containing, family A member 7a |
| chr7_+_15329819 | 3.07 |
ENSDART00000006018
|
mespaa
|
mesoderm posterior aa |
| chr22_+_15315655 | 3.06 |
ENSDART00000141249
|
sult3st3
|
sulfotransferase family 3, cytosolic sulfotransferase 3 |
| chr5_-_28016805 | 3.05 |
ENSDART00000078642
|
vps37b
|
vacuolar protein sorting 37 homolog B (S. cerevisiae) |
| chr24_+_35564668 | 3.04 |
ENSDART00000122734
|
cebpd
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr7_+_15313443 | 3.04 |
ENSDART00000045385
|
mespba
|
mesoderm posterior ba |
| chr23_-_16734009 | 3.03 |
ENSDART00000125449
|
si:ch211-224l10.4
|
si:ch211-224l10.4 |
| chr11_+_21050326 | 3.02 |
ENSDART00000065984
|
zgc:113307
|
zgc:113307 |
| chr25_+_3507368 | 3.01 |
ENSDART00000157777
|
zgc:153293
|
zgc:153293 |
| chr24_-_40700596 | 3.01 |
ENSDART00000162635
|
smyhc2
|
slow myosin heavy chain 2 |
| chr6_-_26895314 | 2.98 |
ENSDART00000134259
|
hdlbpa
|
high density lipoprotein binding protein a |
| chr3_+_47245764 | 2.97 |
ENSDART00000193581
|
tnfsf14
|
TNF superfamily member 14 |
| chr9_+_8380728 | 2.97 |
ENSDART00000133501
|
si:ch1073-75o15.4
|
si:ch1073-75o15.4 |
| chr11_+_37251825 | 2.96 |
ENSDART00000169804
|
il17rc
|
interleukin 17 receptor C |
| chr5_-_16475682 | 2.96 |
ENSDART00000090695
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr9_-_7390388 | 2.94 |
ENSDART00000132392
|
slc23a3
|
solute carrier family 23, member 3 |
| chr5_+_61843752 | 2.94 |
ENSDART00000130940
|
CR759879.1
|
Danio rerio interferon-induced protein with tetratricopeptide repeats 5 (LOC572297), mRNA. |
| chr7_+_25221757 | 2.93 |
ENSDART00000173551
|
exoc6b
|
exocyst complex component 6B |
Network of associatons between targets according to the STRING database.
First level regulatory network of tbr1b_tbx16+tbx16l+tbx2a+tbx2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.2 | GO:0090008 | hypoblast development(GO:0090008) |
| 2.6 | 7.8 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 2.4 | 7.2 | GO:0005986 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 2.3 | 9.1 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 2.2 | 6.7 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 2.1 | 6.4 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 1.6 | 4.9 | GO:0090134 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 1.5 | 1.5 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 1.4 | 8.5 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 1.3 | 4.0 | GO:0052576 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 1.3 | 5.3 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 1.3 | 3.9 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 1.3 | 8.8 | GO:0006953 | acute-phase response(GO:0006953) |
| 1.3 | 3.8 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 1.1 | 4.4 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 1.0 | 13.5 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 1.0 | 3.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 1.0 | 4.9 | GO:0000741 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.9 | 2.8 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.9 | 2.8 | GO:1902103 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.9 | 19.6 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.9 | 6.1 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.8 | 15.1 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.8 | 6.8 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.7 | 11.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.7 | 2.2 | GO:0032640 | tumor necrosis factor production(GO:0032640) tumor necrosis factor superfamily cytokine production(GO:0071706) |
| 0.7 | 2.8 | GO:0055021 | regulation of cardiac muscle tissue growth(GO:0055021) regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.7 | 2.8 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.7 | 2.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.7 | 2.7 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.7 | 10.7 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.7 | 4.0 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.7 | 2.0 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.6 | 2.6 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.6 | 1.9 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.6 | 3.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.6 | 1.2 | GO:0070199 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.6 | 8.0 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.6 | 1.2 | GO:0048322 | axial mesoderm formation(GO:0048320) axial mesodermal cell differentiation(GO:0048321) axial mesodermal cell fate commitment(GO:0048322) axial mesodermal cell fate specification(GO:0048327) |
| 0.6 | 9.0 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.6 | 5.4 | GO:0016121 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.6 | 2.4 | GO:0061033 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.6 | 3.4 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.6 | 2.8 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.5 | 2.2 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 0.5 | 8.0 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.5 | 9.0 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.5 | 3.2 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.5 | 1.6 | GO:0046689 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.5 | 3.7 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.5 | 2.1 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.5 | 5.5 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.5 | 2.0 | GO:0034087 | establishment of mitotic sister chromatid cohesion(GO:0034087) mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.5 | 1.5 | GO:0060584 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.5 | 1.9 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.5 | 1.4 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.5 | 1.4 | GO:1901004 | ubiquinone-6 metabolic process(GO:1901004) ubiquinone-6 biosynthetic process(GO:1901006) |
| 0.5 | 3.6 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.5 | 4.1 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.4 | 1.3 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.4 | 2.7 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.4 | 2.1 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.4 | 1.3 | GO:0001783 | B cell apoptotic process(GO:0001783) regulation of B cell apoptotic process(GO:0002902) regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.4 | 3.4 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.4 | 1.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.4 | 1.6 | GO:0003413 | plasma membrane repair(GO:0001778) chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.4 | 2.0 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.4 | 3.2 | GO:0031179 | peptide modification(GO:0031179) |
| 0.4 | 2.4 | GO:0033986 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.4 | 8.2 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.4 | 6.9 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.4 | 1.5 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.4 | 1.5 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.4 | 1.1 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.4 | 1.1 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.4 | 1.1 | GO:0032369 | negative regulation of lipid transport(GO:0032369) |
| 0.4 | 2.1 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.4 | 2.5 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.4 | 4.2 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.3 | 3.8 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.3 | 0.7 | GO:0002676 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.3 | 6.1 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.3 | 2.4 | GO:0045901 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.3 | 2.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.3 | 1.3 | GO:0070317 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.3 | 1.3 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.3 | 2.6 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.3 | 1.0 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.3 | 5.4 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.3 | 0.9 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.3 | 1.6 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.3 | 9.4 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.3 | 0.9 | GO:1903961 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.3 | 5.1 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.3 | 2.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.3 | 1.4 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.3 | 2.9 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.3 | 1.1 | GO:0015878 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.3 | 1.4 | GO:0071305 | cellular response to vitamin D(GO:0071305) |
| 0.3 | 3.1 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.3 | 2.4 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.3 | 1.9 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.3 | 1.3 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.3 | 2.6 | GO:0086003 | cardiac muscle cell contraction(GO:0086003) |
| 0.3 | 1.8 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.3 | 1.3 | GO:0007584 | response to nutrient(GO:0007584) |
| 0.3 | 6.3 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.3 | 1.8 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.3 | 1.0 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.2 | 10.2 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.2 | 3.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.2 | 2.2 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.2 | 2.7 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.2 | 1.2 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.2 | 3.1 | GO:0003128 | heart field specification(GO:0003128) |
| 0.2 | 5.9 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.2 | 0.9 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.2 | 2.5 | GO:0072698 | protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.2 | 3.4 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.2 | 0.7 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.2 | 2.1 | GO:0021794 | thalamus development(GO:0021794) |
| 0.2 | 3.2 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.2 | 2.3 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.2 | 0.9 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.2 | 1.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.2 | 7.9 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.2 | 11.5 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.2 | 1.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.2 | 1.5 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.2 | 1.5 | GO:0003262 | endocardial progenitor cell migration to the midline involved in heart field formation(GO:0003262) |
| 0.2 | 0.6 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.2 | 1.5 | GO:0046292 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.2 | 2.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 0.8 | GO:0051645 | Golgi localization(GO:0051645) maintenance of centrosome location(GO:0051661) |
| 0.2 | 0.6 | GO:0098725 | somatic stem cell population maintenance(GO:0035019) symmetric cell division(GO:0098725) |
| 0.2 | 1.0 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.2 | 1.0 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.2 | 0.6 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.2 | 1.2 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.2 | 1.2 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.2 | 4.5 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.2 | 1.2 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.2 | 3.1 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.2 | 21.4 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.2 | 1.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.2 | 3.2 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.2 | 1.8 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.2 | 1.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.2 | 0.5 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.2 | 0.5 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.2 | 0.7 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 3.5 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.2 | 4.4 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.2 | 3.1 | GO:0030168 | platelet activation(GO:0030168) |
| 0.2 | 0.7 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.2 | 1.0 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.2 | 4.6 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.2 | 0.3 | GO:0030238 | female sex determination(GO:0030237) male sex determination(GO:0030238) |
| 0.2 | 1.3 | GO:0071459 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.2 | 2.0 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.2 | 2.0 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.2 | 0.3 | GO:0072679 | thymocyte migration(GO:0072679) |
| 0.2 | 7.1 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.2 | 1.0 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.2 | 3.5 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.2 | 1.3 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.2 | 0.9 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.2 | 0.8 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.2 | 1.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.2 | 0.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 2.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.2 | 3.0 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 5.2 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 0.7 | GO:0071265 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.1 | 0.6 | GO:0042304 | regulation of fatty acid biosynthetic process(GO:0042304) |
| 0.1 | 4.1 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 1.6 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 1.3 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 1.7 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 1.6 | GO:0035587 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.1 | 2.8 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 7.3 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.1 | 2.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 1.9 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.1 | 2.9 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.1 | 1.4 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.8 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 | 0.8 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.9 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 2.3 | GO:0018279 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 0.9 | GO:0033628 | regulation of cell adhesion mediated by integrin(GO:0033628) |
| 0.1 | 0.5 | GO:0007220 | Notch receptor processing(GO:0007220) beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.1 | 0.1 | GO:2000257 | regulation of protein activation cascade(GO:2000257) |
| 0.1 | 3.7 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.1 | 0.7 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 1.6 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 0.6 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.7 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.1 | 1.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.6 | GO:0045581 | CD4-positive, alpha-beta T cell differentiation involved in immune response(GO:0002294) T-helper cell lineage commitment(GO:0002295) T cell lineage commitment(GO:0002360) alpha-beta T cell lineage commitment(GO:0002363) T-helper cell differentiation(GO:0042093) CD4-positive or CD8-positive, alpha-beta T cell lineage commitment(GO:0043369) negative regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043371) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) negative regulation of T cell differentiation(GO:0045581) negative regulation of lymphocyte differentiation(GO:0045620) regulation of T-helper cell differentiation(GO:0045622) negative regulation of T-helper cell differentiation(GO:0045623) negative regulation of alpha-beta T cell differentiation(GO:0046639) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) regulation of T-helper 17 type immune response(GO:2000316) negative regulation of T-helper 17 type immune response(GO:2000317) regulation of T-helper 17 cell differentiation(GO:2000319) negative regulation of T-helper 17 cell differentiation(GO:2000320) regulation of T-helper 17 cell lineage commitment(GO:2000328) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.1 | 1.3 | GO:0003207 | cardiac chamber formation(GO:0003207) |
| 0.1 | 1.0 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 1.2 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 3.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 0.3 | GO:0003222 | ventricular trabecula myocardium morphogenesis(GO:0003222) |
| 0.1 | 1.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 1.5 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.1 | 0.9 | GO:0002761 | regulation of myeloid leukocyte differentiation(GO:0002761) |
| 0.1 | 1.3 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.1 | 1.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 5.7 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.1 | 2.1 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 2.7 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 3.1 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.1 | 1.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 2.8 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 2.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.9 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 2.3 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 0.4 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.1 | 0.4 | GO:0048901 | anterior lateral line neuromast development(GO:0048901) anterior lateral line neuromast hair cell differentiation(GO:0048903) |
| 0.1 | 0.6 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.1 | 3.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 1.3 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.1 | 0.5 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 3.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.6 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 2.8 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 1.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 1.1 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 2.1 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.1 | 3.6 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.1 | 4.1 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.1 | 4.5 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.1 | 0.9 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 1.7 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 4.0 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.1 | 1.9 | GO:0090148 | membrane fission(GO:0090148) |
| 0.1 | 2.9 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 1.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 1.1 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 2.9 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.1 | 1.2 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.1 | 1.4 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.1 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.7 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 6.5 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.1 | 5.6 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.1 | 3.6 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.1 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 3.6 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 1.0 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.1 | 11.0 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.1 | 3.4 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.1 | 1.2 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 0.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.9 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 0.3 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 0.9 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 | 0.6 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 3.6 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.1 | 1.5 | GO:0042113 | B cell activation(GO:0042113) |
| 0.1 | 4.6 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.1 | 4.6 | GO:0001894 | tissue homeostasis(GO:0001894) |
| 0.1 | 13.6 | GO:0009617 | response to bacterium(GO:0009617) |
| 0.1 | 2.3 | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation(GO:0050731) |
| 0.1 | 0.9 | GO:0071545 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.1 | 2.5 | GO:0051262 | protein tetramerization(GO:0051262) |
| 0.1 | 1.0 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.7 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.1 | 2.3 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.1 | 1.3 | GO:0048546 | digestive tract morphogenesis(GO:0048546) |
| 0.1 | 0.4 | GO:0000272 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 0.6 | GO:0002695 | negative regulation of leukocyte activation(GO:0002695) negative regulation of cell activation(GO:0050866) negative regulation of lymphocyte activation(GO:0051250) |
| 0.1 | 0.9 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 1.4 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.1 | 0.6 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.1 | 3.6 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.1 | 2.9 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.1 | 1.5 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 0.8 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 2.8 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 0.6 | GO:0043651 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.4 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 1.6 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.1 | 0.3 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.1 | 2.4 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.6 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.0 | 3.4 | GO:0009612 | response to mechanical stimulus(GO:0009612) |
| 0.0 | 0.7 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.4 | GO:0001707 | mesoderm formation(GO:0001707) |
| 0.0 | 3.3 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.5 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 5.2 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.0 | 1.3 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 1.3 | GO:0006956 | complement activation(GO:0006956) |
| 0.0 | 1.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.7 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 1.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.2 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 2.6 | GO:0033334 | fin morphogenesis(GO:0033334) |
| 0.0 | 0.6 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 1.9 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 2.7 | GO:0009308 | amine metabolic process(GO:0009308) |
| 0.0 | 2.8 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.0 | 0.5 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.6 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.7 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 1.6 | GO:0008589 | regulation of smoothened signaling pathway(GO:0008589) |
| 0.0 | 3.8 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 3.2 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.8 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.0 | 0.5 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 2.0 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.6 | GO:0033522 | histone H2A ubiquitination(GO:0033522) |
| 0.0 | 1.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 1.2 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
| 0.0 | 0.7 | GO:0051283 | sequestering of calcium ion(GO:0051208) release of sequestered calcium ion into cytosol(GO:0051209) sequestering of metal ion(GO:0051238) regulation of sequestering of calcium ion(GO:0051282) negative regulation of sequestering of calcium ion(GO:0051283) |
| 0.0 | 1.6 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 1.1 | GO:0001946 | lymphangiogenesis(GO:0001946) lymph vessel morphogenesis(GO:0036303) |
| 0.0 | 1.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.6 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0009182 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.6 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:0090151 | protein import into mitochondrial inner membrane(GO:0045039) establishment of protein localization to mitochondrial membrane(GO:0090151) |
| 0.0 | 0.5 | GO:1901661 | quinone metabolic process(GO:1901661) |
| 0.0 | 0.4 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.5 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.7 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.1 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.5 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.3 | GO:0043270 | positive regulation of ion transmembrane transporter activity(GO:0032414) positive regulation of transmembrane transport(GO:0034764) positive regulation of ion transmembrane transport(GO:0034767) positive regulation of ion transport(GO:0043270) positive regulation of cation transmembrane transport(GO:1904064) |
| 0.0 | 0.1 | GO:0003315 | heart rudiment formation(GO:0003315) |
| 0.0 | 0.6 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.2 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.7 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.7 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 1.1 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.7 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.2 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 1.5 | GO:0030155 | regulation of cell adhesion(GO:0030155) |
| 0.0 | 0.3 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.7 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.5 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.6 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 1.1 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.9 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 0.1 | GO:0036372 | opsin transport(GO:0036372) |
| 0.0 | 2.6 | GO:0006955 | immune response(GO:0006955) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.3 | GO:1990923 | PET complex(GO:1990923) |
| 0.8 | 3.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.7 | 2.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.7 | 3.4 | GO:0097433 | dense body(GO:0097433) |
| 0.6 | 3.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.6 | 1.8 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.6 | 10.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.5 | 9.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.5 | 3.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.4 | 1.3 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.4 | 2.0 | GO:0031415 | NatA complex(GO:0031415) |
| 0.4 | 4.0 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.4 | 2.1 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.3 | 1.4 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.3 | 10.5 | GO:0031672 | A band(GO:0031672) |
| 0.3 | 18.3 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.3 | 1.1 | GO:0031511 | Mis6-Sim4 complex(GO:0031511) |
| 0.3 | 2.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 4.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.3 | 1.3 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.2 | 11.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 1.0 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.2 | 0.9 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.2 | 3.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 6.2 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.2 | 9.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 0.8 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.2 | 1.4 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 1.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.2 | 2.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 0.9 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.2 | 1.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.2 | 1.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.2 | 3.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 10.5 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.2 | 8.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.2 | 3.2 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.2 | 3.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.2 | 2.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 2.6 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.2 | 1.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 0.6 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.1 | 0.4 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 0.1 | 0.9 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 1.9 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 15.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 2.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 3.1 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 1.8 | GO:0000780 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.1 | 0.6 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.5 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 6.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 42.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 0.8 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 4.4 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) |
| 0.1 | 4.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 2.8 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 3.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.8 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 1.0 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 0.8 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 2.1 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 3.2 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 2.4 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 1.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 9.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 0.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.9 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 91.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 4.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.6 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 1.2 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 1.1 | GO:0098798 | mitochondrial protein complex(GO:0098798) |
| 0.1 | 7.7 | GO:0030055 | cell-substrate junction(GO:0030055) |
| 0.1 | 1.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 7.5 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 4.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.2 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 12.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.9 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 4.1 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 2.2 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 0.7 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 6.8 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 0.5 | GO:0035060 | brahma complex(GO:0035060) |
| 0.1 | 1.1 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 3.9 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 0.9 | GO:0044545 | NSL complex(GO:0044545) |
| 0.1 | 0.3 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 0.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 1.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 1.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 4.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 0.5 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 2.0 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.9 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 2.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 2.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.7 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.0 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 2.5 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 1.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.7 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 1.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 1.6 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 1.3 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 7.4 | GO:0005740 | mitochondrial envelope(GO:0005740) |
| 0.0 | 4.3 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 9.3 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.1 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 2.1 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.8 | GO:0005814 | centriole(GO:0005814) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 15.9 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 3.0 | 9.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 2.4 | 7.2 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 2.0 | 7.9 | GO:0070052 | collagen V binding(GO:0070052) |
| 1.8 | 7.0 | GO:0015105 | arsenite transmembrane transporter activity(GO:0015105) |
| 1.3 | 4.0 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 1.3 | 4.0 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 1.3 | 3.9 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 1.2 | 8.5 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 1.2 | 3.5 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 1.0 | 13.5 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 1.0 | 13.4 | GO:0070697 | activin receptor binding(GO:0070697) |
| 1.0 | 3.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.9 | 2.8 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.9 | 6.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.8 | 5.8 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.8 | 3.3 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.8 | 12.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.8 | 4.0 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.8 | 3.8 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.8 | 8.3 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.7 | 4.3 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.7 | 3.4 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.7 | 5.3 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.6 | 6.2 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.6 | 5.4 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.6 | 2.3 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.6 | 1.7 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.6 | 6.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.6 | 3.4 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.5 | 3.8 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.5 | 2.7 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.5 | 2.1 | GO:0051916 | C-X-C chemokine receptor activity(GO:0016494) granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.5 | 9.0 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.5 | 7.9 | GO:0031386 | protein tag(GO:0031386) |
| 0.5 | 2.9 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.5 | 2.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.5 | 2.8 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.5 | 3.6 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.4 | 2.7 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.4 | 1.7 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.4 | 5.5 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.4 | 13.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.4 | 6.1 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.4 | 2.8 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.4 | 4.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.4 | 3.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.4 | 6.6 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.4 | 3.1 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.4 | 4.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.4 | 3.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.4 | 4.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.4 | 1.5 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.4 | 3.4 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.4 | 1.1 | GO:0047777 | (3S)-citramalyl-CoA lyase activity(GO:0047777) |
| 0.4 | 1.5 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.4 | 1.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.4 | 3.2 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.3 | 1.7 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.3 | 4.7 | GO:0002020 | protease binding(GO:0002020) |
| 0.3 | 1.3 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.3 | 3.3 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.3 | 2.9 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.3 | 2.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 0.6 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.3 | 1.2 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.3 | 1.1 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.3 | 1.1 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.3 | 1.3 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.3 | 1.3 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.3 | 7.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.3 | 0.8 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 0.3 | 3.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.3 | 1.3 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.3 | 3.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.2 | 11.2 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.2 | 1.7 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.2 | 2.7 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 0.7 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.2 | 2.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.2 | 2.6 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.2 | 12.8 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.2 | 3.5 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.2 | 0.7 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.2 | 1.6 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.2 | 0.4 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.2 | 1.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 3.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 2.2 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 0.9 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.2 | 3.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 2.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 6.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 1.2 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.2 | 2.0 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.2 | 3.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.2 | 0.5 | GO:0071077 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.2 | 6.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.2 | 1.3 | GO:0043394 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.2 | 0.6 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.2 | 2.4 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.2 | 0.8 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.2 | 2.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 2.8 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 3.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 3.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.3 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 1.7 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.4 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) |
| 0.1 | 1.0 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.1 | 0.6 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.1 | 34.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 1.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 2.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 1.9 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 1.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.6 | GO:1902388 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.1 | 2.0 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 5.9 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 0.8 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 2.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.9 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.9 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 3.0 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.3 | GO:0030251 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.1 | 25.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 1.6 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 5.7 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 0.7 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.1 | 0.8 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 3.2 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.1 | 1.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 1.2 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 2.1 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 7.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 1.8 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 1.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 1.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 3.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 1.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 3.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.4 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.1 | 18.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 0.4 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.1 | 0.6 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 0.7 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 2.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 1.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 2.1 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.1 | 1.1 | GO:0005123 | death receptor binding(GO:0005123) nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.1 | 8.7 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 0.9 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 11.9 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 4.4 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.1 | 0.4 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 11.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 2.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.6 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 0.8 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 0.7 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.1 | 1.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.7 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.1 | 1.3 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 0.9 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 1.1 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 1.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.9 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 0.3 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 1.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 0.5 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 1.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 0.5 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 0.9 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.7 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 1.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.5 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 5.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 0.6 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 0.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.7 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 1.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.3 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.1 | 2.1 | GO:0005126 | cytokine receptor binding(GO:0005126) |
| 0.1 | 1.7 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 1.0 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0005461 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.0 | 2.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 29.4 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 3.1 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 3.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.4 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.8 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 17.4 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 1.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.9 | GO:0099604 | ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.2 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.9 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.0 | 1.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.7 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.9 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 1.1 | GO:0004532 | 3'-5'-exoribonuclease activity(GO:0000175) exoribonuclease activity(GO:0004532) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.0 | 0.4 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 4.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 1.9 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.4 | GO:0005217 | intracellular ligand-gated ion channel activity(GO:0005217) intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 2.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 4.7 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 2.1 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 1.5 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 1.3 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.2 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) phosphoprotein binding(GO:0051219) |
| 0.0 | 1.1 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 1.2 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 3.3 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 0.3 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 11.2 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.1 | GO:0070035 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 0.1 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.0 | 0.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 1.3 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.3 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 2.6 | GO:0008194 | UDP-glycosyltransferase activity(GO:0008194) |
| 0.0 | 0.1 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.0 | GO:0001164 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.2 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 1.0 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.8 | 7.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.6 | 7.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.5 | 0.9 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.5 | 9.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.4 | 18.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.4 | 0.9 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.4 | 6.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.4 | 10.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.4 | 3.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.4 | 15.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.3 | 2.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 2.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.2 | 3.9 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.2 | 1.0 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 7.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 2.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.2 | 1.8 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.2 | 4.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 3.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.2 | 0.9 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 1.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 2.8 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 1.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 7.1 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 2.6 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.1 | 3.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 0.9 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 4.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 1.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 14.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.0 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 4.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 0.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 6.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 1.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 1.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 0.8 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 1.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 0.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 0.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.1 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 2.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 6.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 6.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.6 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 1.2 | 9.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.9 | 9.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.7 | 5.8 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.6 | 7.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.5 | 3.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.4 | 4.0 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.4 | 1.3 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.4 | 2.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.4 | 4.7 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.3 | 2.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.3 | 1.9 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.3 | 3.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 3.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.3 | 3.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.3 | 3.8 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.2 | 6.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.2 | 2.5 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.2 | 2.4 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.2 | 2.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 0.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 4.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 11.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.2 | 1.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.2 | 3.0 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 4.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 2.5 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.2 | 6.4 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.2 | 7.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 11.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.2 | 7.9 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.2 | 3.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 0.7 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.1 | 5.4 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 2.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 0.9 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 0.7 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 2.2 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 0.8 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.1 | 1.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 3.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.0 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 1.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 2.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.6 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 0.5 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 1.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 4.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 0.5 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.1 | 2.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 3.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 2.9 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 1.2 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 1.2 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.9 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.4 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.3 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.3 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 1.6 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.3 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.4 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |