Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
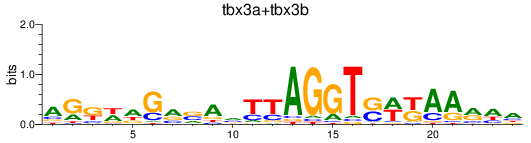
Results for tbx3a+tbx3b
Z-value: 0.94
Transcription factors associated with tbx3a+tbx3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tbx3a
|
ENSDARG00000002216 | T-box transcription factor 3a |
|
tbx3b
|
ENSDARG00000061509 | T-box transcription factor 3b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tbx3a | dr11_v1_chr5_-_72324371_72324371 | -0.51 | 1.5e-07 | Click! |
| tbx3b | dr11_v1_chr5_+_26913120_26913120 | -0.22 | 3.7e-02 | Click! |
Activity profile of tbx3a+tbx3b motif
Sorted Z-values of tbx3a+tbx3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_5547185 | 17.85 |
ENSDART00000193977
|
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr25_-_13188678 | 13.43 |
ENSDART00000125754
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr7_-_73752955 | 12.45 |
ENSDART00000171254
ENSDART00000009888 |
casq1b
|
calsequestrin 1b |
| chr13_-_37180815 | 12.21 |
ENSDART00000139907
|
si:dkeyp-77c8.1
|
si:dkeyp-77c8.1 |
| chr23_-_5683147 | 11.71 |
ENSDART00000102766
ENSDART00000067351 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr13_-_51903150 | 7.14 |
ENSDART00000090644
|
mrpl2
|
mitochondrial ribosomal protein L2 |
| chr12_-_26415499 | 6.82 |
ENSDART00000185779
|
synpo2lb
|
synaptopodin 2-like b |
| chr18_+_29145681 | 6.69 |
ENSDART00000089031
ENSDART00000193336 |
ppfibp2a
|
PTPRF interacting protein, binding protein 2a (liprin beta 2) |
| chr12_-_26406323 | 5.97 |
ENSDART00000131896
|
myoz1b
|
myozenin 1b |
| chr25_-_27819838 | 5.74 |
ENSDART00000067106
|
lmod2a
|
leiomodin 2 (cardiac) a |
| chr20_+_53577502 | 5.56 |
ENSDART00000126983
|
myh6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr24_+_35947077 | 5.26 |
ENSDART00000173406
|
obscnb
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF b |
| chr24_+_854877 | 5.10 |
ENSDART00000188408
|
piezo2a.2
|
piezo-type mechanosensitive ion channel component 2a, tandem duplicate 2 |
| chr8_+_26818446 | 5.02 |
ENSDART00000134987
ENSDART00000138835 |
si:ch211-156j16.1
|
si:ch211-156j16.1 |
| chr3_-_33967767 | 4.87 |
ENSDART00000151493
ENSDART00000151160 |
ighv1-4
|
immunoglobulin heavy variable 1-4 |
| chr13_+_25486608 | 4.81 |
ENSDART00000057689
|
bag3
|
BCL2 associated athanogene 3 |
| chr1_-_38813679 | 4.78 |
ENSDART00000148917
|
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr9_-_105135 | 4.78 |
ENSDART00000180126
|
FQ377903.3
|
|
| chr2_-_37103622 | 4.34 |
ENSDART00000137849
|
zgc:101744
|
zgc:101744 |
| chr21_-_33478164 | 4.20 |
ENSDART00000191542
|
si:ch73-42p12.2
|
si:ch73-42p12.2 |
| chr23_+_2421689 | 4.18 |
ENSDART00000180200
|
tcp1
|
t-complex 1 |
| chr17_+_8175998 | 4.09 |
ENSDART00000131200
|
myct1b
|
myc target 1b |
| chr6_-_1514767 | 3.95 |
ENSDART00000067586
|
chchd6b
|
coiled-coil-helix-coiled-coil-helix domain containing 6b |
| chr8_-_25336589 | 3.32 |
ENSDART00000009682
|
atp5pb
|
ATP synthase peripheral stalk-membrane subunit b |
| chr3_+_5297493 | 3.30 |
ENSDART00000138596
|
si:ch211-150d5.3
|
si:ch211-150d5.3 |
| chr21_+_21655755 | 3.28 |
ENSDART00000079524
|
or125-8
|
odorant receptor, family E, subfamily 125, member 8 |
| chr5_-_38197080 | 3.19 |
ENSDART00000140708
|
si:ch211-284e13.9
|
si:ch211-284e13.9 |
| chr18_+_37272568 | 3.13 |
ENSDART00000132749
|
tmem123
|
transmembrane protein 123 |
| chr3_-_1187727 | 2.96 |
ENSDART00000180343
|
smdt1a
|
single-pass membrane protein with aspartate-rich tail 1a |
| chr17_-_50071748 | 2.95 |
ENSDART00000075188
|
zgc:113886
|
zgc:113886 |
| chr17_-_47090440 | 2.90 |
ENSDART00000163542
|
CABZ01056321.1
|
|
| chr1_+_218524 | 2.87 |
ENSDART00000109529
|
tmco3
|
transmembrane and coiled-coil domains 3 |
| chr24_-_31306724 | 2.83 |
ENSDART00000165399
|
acp5b
|
acid phosphatase 5b, tartrate resistant |
| chr22_+_344763 | 2.67 |
ENSDART00000181934
|
CU914780.1
|
|
| chr1_+_56502706 | 2.57 |
ENSDART00000188665
|
CABZ01059403.1
|
|
| chr12_+_27141140 | 2.57 |
ENSDART00000136415
|
hoxb1b
|
homeobox B1b |
| chr3_-_6191950 | 2.56 |
ENSDART00000161931
|
si:ch73-144l3.1
|
si:ch73-144l3.1 |
| chr7_-_3836635 | 2.55 |
ENSDART00000104350
|
si:dkey-88n24.8
|
si:dkey-88n24.8 |
| chr7_+_65121743 | 2.50 |
ENSDART00000108471
|
ipo7
|
importin 7 |
| chr5_+_29803702 | 2.49 |
ENSDART00000147569
|
usf1l
|
upstream transcription factor 1, like |
| chr19_+_9212031 | 2.43 |
ENSDART00000052930
|
ndufv1
|
NADH dehydrogenase (ubiquinone) flavoprotein 1 |
| chr19_-_18127808 | 2.43 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr19_-_18127629 | 2.42 |
ENSDART00000187722
|
snx10a
|
sorting nexin 10a |
| chr9_+_50001746 | 2.42 |
ENSDART00000058892
|
slc38a11
|
solute carrier family 38, member 11 |
| chr8_-_32894203 | 2.37 |
ENSDART00000147822
|
si:dkey-56i24.1
|
si:dkey-56i24.1 |
| chr20_+_51464670 | 2.35 |
ENSDART00000150110
|
thbd
|
thrombomodulin |
| chr23_-_20345473 | 2.34 |
ENSDART00000140935
|
si:rp71-17i16.6
|
si:rp71-17i16.6 |
| chr1_-_45177373 | 2.33 |
ENSDART00000143142
ENSDART00000034549 |
zgc:111983
|
zgc:111983 |
| chr7_+_22702437 | 2.33 |
ENSDART00000182054
|
si:dkey-165a24.9
|
si:dkey-165a24.9 |
| chr22_+_9294336 | 2.28 |
ENSDART00000193694
|
si:ch211-250k18.7
|
si:ch211-250k18.7 |
| chr24_-_26479841 | 2.27 |
ENSDART00000079984
|
rpl22l1
|
ribosomal protein L22-like 1 |
| chr5_-_67783593 | 2.26 |
ENSDART00000010934
|
casr
|
calcium-sensing receptor |
| chr14_-_28052474 | 2.23 |
ENSDART00000172948
ENSDART00000135337 |
TSC22D3 (1 of many)
zgc:64189
|
si:ch211-220e11.3 zgc:64189 |
| chr18_-_21748966 | 2.22 |
ENSDART00000181910
|
pskh1
|
protein serine kinase H1 |
| chr12_+_20506197 | 2.22 |
ENSDART00000153010
|
si:zfos-754c12.2
|
si:zfos-754c12.2 |
| chr19_-_23674904 | 2.19 |
ENSDART00000187300
|
BX927234.2
|
|
| chr15_-_21678634 | 2.19 |
ENSDART00000061139
|
bco2b
|
beta-carotene oxygenase 2b |
| chr10_-_15048781 | 2.09 |
ENSDART00000038401
ENSDART00000155674 |
si:ch211-95j8.2
|
si:ch211-95j8.2 |
| chr22_-_9157364 | 2.08 |
ENSDART00000182762
|
si:ch211-213a13.5
|
si:ch211-213a13.5 |
| chr13_-_11984867 | 2.05 |
ENSDART00000157538
|
npm3
|
nucleophosmin/nucleoplasmin, 3 |
| chr5_+_29803380 | 2.05 |
ENSDART00000005263
ENSDART00000137558 ENSDART00000146963 ENSDART00000134900 |
usf1l
|
upstream transcription factor 1, like |
| chr16_-_14587332 | 2.04 |
ENSDART00000012479
|
dscc1
|
DNA replication and sister chromatid cohesion 1 |
| chr7_+_2236317 | 2.03 |
ENSDART00000075859
|
zgc:172065
|
zgc:172065 |
| chr3_-_2072630 | 1.97 |
ENSDART00000189404
|
BX005442.3
|
|
| chr8_+_20918207 | 1.96 |
ENSDART00000144039
|
si:ch73-196i15.5
|
si:ch73-196i15.5 |
| chr22_-_9649627 | 1.94 |
ENSDART00000164721
|
si:dkey-286j17.4
|
si:dkey-286j17.4 |
| chr7_-_3826794 | 1.92 |
ENSDART00000064229
ENSDART00000142138 |
si:dkey-88n24.10
|
si:dkey-88n24.10 |
| chr1_-_2453174 | 1.90 |
ENSDART00000055779
ENSDART00000152555 |
ggact.2
ggact.3
|
gamma-glutamylamine cyclotransferase, tandem duplicate 2 gamma-glutamylamine cyclotransferase, tandem duplicate 3 |
| chr16_-_41990421 | 1.81 |
ENSDART00000055921
|
pycard
|
PYD and CARD domain containing |
| chr14_-_8903435 | 1.77 |
ENSDART00000160584
|
zgc:153681
|
zgc:153681 |
| chr24_+_12074340 | 1.76 |
ENSDART00000170997
ENSDART00000166227 |
ccr9b
|
chemokine (C-C motif) receptor 9b |
| chr6_+_15250672 | 1.76 |
ENSDART00000155951
|
si:ch73-23l24.1
|
si:ch73-23l24.1 |
| chr8_-_46897734 | 1.75 |
ENSDART00000138125
|
hes2.2
|
hes family bHLH transcription factor 2, tandem duplicate 2 |
| chr19_+_22930177 | 1.73 |
ENSDART00000151737
|
si:ch211-244a23.1
|
si:ch211-244a23.1 |
| chr14_+_46287296 | 1.72 |
ENSDART00000183620
|
cabp2b
|
calcium binding protein 2b |
| chr4_+_14981854 | 1.70 |
ENSDART00000067046
|
cax1
|
cation/H+ exchanger protein 1 |
| chr12_+_18663154 | 1.69 |
ENSDART00000057918
|
si:ch211-147h1.4
|
si:ch211-147h1.4 |
| chr14_+_20911310 | 1.68 |
ENSDART00000160318
|
lygl2
|
lysozyme g-like 2 |
| chr7_+_22702225 | 1.68 |
ENSDART00000173672
|
si:dkey-165a24.9
|
si:dkey-165a24.9 |
| chr7_-_3836896 | 1.67 |
ENSDART00000136227
|
si:dkey-88n24.8
|
si:dkey-88n24.8 |
| chr5_-_5035683 | 1.62 |
ENSDART00000170025
|
lmx1ba
|
LIM homeobox transcription factor 1, beta a |
| chr24_-_25184553 | 1.61 |
ENSDART00000166917
|
plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr15_-_1765098 | 1.60 |
ENSDART00000149980
ENSDART00000093074 |
bud23
|
BUD23, rRNA methyltransferase and ribosome maturation factor |
| chr4_-_75812937 | 1.57 |
ENSDART00000125096
|
si:ch211-203c5.3
|
si:ch211-203c5.3 |
| chr21_-_20725853 | 1.55 |
ENSDART00000114502
|
si:ch211-22d5.2
|
si:ch211-22d5.2 |
| chr4_-_36792144 | 1.53 |
ENSDART00000159147
|
si:dkeyp-87d1.1
|
si:dkeyp-87d1.1 |
| chr25_-_15512819 | 1.51 |
ENSDART00000142684
|
si:dkeyp-67e1.2
|
si:dkeyp-67e1.2 |
| chr25_-_15504559 | 1.50 |
ENSDART00000139294
|
BX323543.5
|
|
| chr18_-_40913294 | 1.46 |
ENSDART00000059196
ENSDART00000098878 |
polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chr16_-_50918908 | 1.43 |
ENSDART00000108744
|
CU855821.1
|
|
| chr2_-_21438492 | 1.41 |
ENSDART00000046098
|
plcd1b
|
phospholipase C, delta 1b |
| chr2_-_38992304 | 1.40 |
ENSDART00000114085
ENSDART00000146812 |
si:ch211-119o8.6
|
si:ch211-119o8.6 |
| chr3_+_37790351 | 1.39 |
ENSDART00000151506
|
si:dkey-260c8.8
|
si:dkey-260c8.8 |
| chr22_+_15323930 | 1.38 |
ENSDART00000142416
|
si:dkey-236e20.3
|
si:dkey-236e20.3 |
| chr23_+_20687340 | 1.35 |
ENSDART00000143503
|
usp21
|
ubiquitin specific peptidase 21 |
| chr13_+_33462232 | 1.35 |
ENSDART00000177841
|
zgc:136302
|
zgc:136302 |
| chr8_+_23521974 | 1.32 |
ENSDART00000188130
ENSDART00000129378 |
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr6_-_55399214 | 1.31 |
ENSDART00000168367
|
ctsa
|
cathepsin A |
| chr12_+_31537604 | 1.28 |
ENSDART00000153340
|
si:ch73-205h11.1
|
si:ch73-205h11.1 |
| chr4_+_62598975 | 1.26 |
ENSDART00000163548
|
si:ch211-79g12.2
|
si:ch211-79g12.2 |
| chr2_+_20472150 | 1.26 |
ENSDART00000168537
|
agla
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase a |
| chr20_-_34164278 | 1.24 |
ENSDART00000153128
|
hmcn1
|
hemicentin 1 |
| chr3_-_36690348 | 1.21 |
ENSDART00000192513
|
myh11b
|
myosin, heavy chain 11b, smooth muscle |
| chr24_-_23942722 | 1.18 |
ENSDART00000080810
|
arxa
|
aristaless related homeobox a |
| chr5_+_69856153 | 1.17 |
ENSDART00000124128
ENSDART00000073663 |
ugt2a6
|
UDP glucuronosyltransferase 2 family, polypeptide A6 |
| chr4_+_53246788 | 1.17 |
ENSDART00000184708
ENSDART00000169256 |
si:dkey-250k10.1
|
si:dkey-250k10.1 |
| chr14_+_45645024 | 1.16 |
ENSDART00000168278
|
si:ch211-276i12.4
|
si:ch211-276i12.4 |
| chr4_+_77681389 | 1.13 |
ENSDART00000099727
|
gbp4
|
guanylate binding protein 4 |
| chr2_+_27403300 | 1.12 |
ENSDART00000099180
|
elovl8a
|
ELOVL fatty acid elongase 8a |
| chr25_-_35599887 | 1.11 |
ENSDART00000153827
|
clpxb
|
caseinolytic mitochondrial matrix peptidase chaperone subunit b |
| chr4_-_49987980 | 1.09 |
ENSDART00000150428
|
si:dkey-156k2.4
|
si:dkey-156k2.4 |
| chr16_+_32029090 | 1.08 |
ENSDART00000041054
|
tmc4
|
transmembrane channel-like 4 |
| chr5_-_10236599 | 1.07 |
ENSDART00000099834
|
si:ch73-42k18.1
|
si:ch73-42k18.1 |
| chr25_-_3647277 | 0.99 |
ENSDART00000166363
|
si:ch211-272n13.3
|
si:ch211-272n13.3 |
| chr17_-_53329704 | 0.99 |
ENSDART00000193895
|
exd1
|
exonuclease 3'-5' domain containing 1 |
| chr21_+_7332963 | 0.97 |
ENSDART00000169834
|
AP3B1
|
adaptor related protein complex 3 beta 1 subunit |
| chr20_-_26532167 | 0.95 |
ENSDART00000061914
|
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr3_+_17314997 | 0.92 |
ENSDART00000139763
|
stat5a
|
signal transducer and activator of transcription 5a |
| chr15_-_41677689 | 0.92 |
ENSDART00000187063
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr7_+_13756374 | 0.90 |
ENSDART00000180808
|
rasl12
|
RAS-like, family 12 |
| chr15_-_35031523 | 0.89 |
ENSDART00000154848
|
si:ch211-272b8.6
|
si:ch211-272b8.6 |
| chr21_+_21611867 | 0.87 |
ENSDART00000189148
|
b9d2
|
B9 domain containing 2 |
| chr9_-_25094181 | 0.86 |
ENSDART00000132160
|
rubcnl
|
RUN and cysteine rich domain containing beclin 1 interacting protein like |
| chr21_-_27195256 | 0.85 |
ENSDART00000133152
ENSDART00000065401 |
zgc:110782
|
zgc:110782 |
| chr25_-_15496485 | 0.81 |
ENSDART00000140245
|
si:dkeyp-67e1.6
|
si:dkeyp-67e1.6 |
| chr8_+_15251448 | 0.78 |
ENSDART00000063717
|
zgc:171480
|
zgc:171480 |
| chr13_-_32635859 | 0.78 |
ENSDART00000146249
ENSDART00000145395 ENSDART00000148040 ENSDART00000100650 |
matn3b
|
matrilin 3b |
| chr11_-_23219367 | 0.75 |
ENSDART00000003646
|
optc
|
opticin |
| chr9_-_374693 | 0.74 |
ENSDART00000166571
|
si:dkey-11f4.7
|
si:dkey-11f4.7 |
| chr13_-_32577386 | 0.66 |
ENSDART00000016535
|
kcns3a
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3a |
| chr2_+_42005475 | 0.63 |
ENSDART00000056461
|
gbp2
|
guanylate binding protein 2 |
| chr16_+_23331633 | 0.59 |
ENSDART00000187455
|
krtcap2
|
keratinocyte associated protein 2 |
| chr2_-_43745146 | 0.58 |
ENSDART00000056164
ENSDART00000098052 |
ftr96
|
finTRIM family, member 96 |
| chr22_-_5441893 | 0.56 |
ENSDART00000161421
|
zgc:194627
|
zgc:194627 |
| chr21_+_21612214 | 0.51 |
ENSDART00000008099
|
b9d2
|
B9 domain containing 2 |
| chr12_+_20583552 | 0.49 |
ENSDART00000170035
|
arsg
|
arylsulfatase G |
| chr19_-_25464291 | 0.48 |
ENSDART00000112915
|
umad1
|
UBAP1-MVB12-associated (UMA) domain containing 1 |
| chr9_-_23156908 | 0.44 |
ENSDART00000135461
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr12_-_26491464 | 0.39 |
ENSDART00000153361
|
si:dkey-287g12.6
|
si:dkey-287g12.6 |
| chr2_+_43895103 | 0.35 |
ENSDART00000007074
|
gbp3
|
guanylate binding protein 3 |
| chr8_-_50979047 | 0.33 |
ENSDART00000184788
ENSDART00000180906 |
zgc:91909
|
zgc:91909 |
| chr15_-_38154616 | 0.30 |
ENSDART00000099392
|
irgq2
|
immunity-related GTPase family, q2 |
| chr4_+_77908076 | 0.26 |
ENSDART00000168811
|
si:zfos-2131b9.2
|
si:zfos-2131b9.2 |
| chr13_+_3954715 | 0.21 |
ENSDART00000182477
ENSDART00000192142 ENSDART00000190962 |
lrrc73
|
leucine rich repeat containing 73 |
| chr4_+_43700319 | 0.18 |
ENSDART00000141967
|
si:ch211-226o13.1
|
si:ch211-226o13.1 |
| chr7_+_17106160 | 0.12 |
ENSDART00000190048
ENSDART00000180004 ENSDART00000013409 |
prmt3
|
protein arginine methyltransferase 3 |
| chr7_-_33960170 | 0.08 |
ENSDART00000180766
|
skor1a
|
SKI family transcriptional corepressor 1a |
| chr13_+_30163515 | 0.06 |
ENSDART00000040926
|
eif4ebp2
|
eukaryotic translation initiation factor 4E binding protein 2 |
| chr23_-_19831739 | 0.04 |
ENSDART00000125066
|
haus7
|
HAUS augmin-like complex, subunit 7 |
| chr23_-_15090782 | 0.03 |
ENSDART00000133624
|
si:ch211-218g4.2
|
si:ch211-218g4.2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of tbx3a+tbx3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 17.9 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 4.2 | 12.5 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 2.9 | 11.7 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 1.6 | 4.8 | GO:0010660 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 1.0 | 4.8 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.9 | 5.6 | GO:0055014 | atrial cardiac muscle cell development(GO:0055014) |
| 0.9 | 2.6 | GO:0021570 | rhombomere 4 development(GO:0021570) rhombomere 4 morphogenesis(GO:0021661) |
| 0.7 | 2.0 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.6 | 5.8 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.6 | 5.7 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.6 | 1.7 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.4 | 1.6 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.4 | 2.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.3 | 3.2 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.3 | 1.2 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.3 | 2.2 | GO:0016121 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.3 | 2.8 | GO:0045453 | bone resorption(GO:0045453) |
| 0.2 | 6.8 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.2 | 1.3 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.2 | 1.5 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.2 | 7.1 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.2 | 0.6 | GO:0042543 | protein N-linked glycosylation via arginine(GO:0042543) |
| 0.2 | 1.1 | GO:0044364 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.2 | 3.0 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 | 2.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 | 0.9 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.1 | 1.6 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 3.9 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.1 | 3.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 2.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 1.3 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 4.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 6.7 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 1.2 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.1 | 0.9 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 1.4 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.1 | 1.8 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.1 | 1.9 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.1 | 1.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.0 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 4.8 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 4.2 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 1.5 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 5.3 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 1.2 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 2.3 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 1.4 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 1.8 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 1.4 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 6.2 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 1.0 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 4.2 | GO:0060271 | cilium morphogenesis(GO:0060271) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 12.5 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.7 | 2.0 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.5 | 1.8 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.4 | 4.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.4 | 3.9 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.3 | 17.5 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.3 | 5.3 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 3.0 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 3.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.2 | 1.0 | GO:1990923 | PET complex(GO:1990923) |
| 0.2 | 17.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.2 | 7.9 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 1.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 4.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 1.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 2.4 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 6.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 3.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 2.3 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 1.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 17.9 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 1.1 | 11.7 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.6 | 6.0 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.5 | 5.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.5 | 4.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.4 | 1.7 | GO:0015369 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.4 | 1.7 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.4 | 2.8 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.3 | 1.3 | GO:0004135 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.3 | 1.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 2.2 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.2 | 6.9 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 2.0 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 2.9 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 0.9 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 1.8 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 2.4 | GO:0010181 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) FMN binding(GO:0010181) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.6 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 4.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 3.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.3 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 0.4 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 5.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 1.5 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 1.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 10.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 1.4 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 3.3 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 6.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 5.3 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.5 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.0 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 1.6 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 1.4 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 5.9 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 6.5 | GO:0005525 | GTP binding(GO:0005525) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 2.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 2.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.3 | 2.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 4.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.2 | 5.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 2.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.5 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 0.9 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.8 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 2.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |