Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
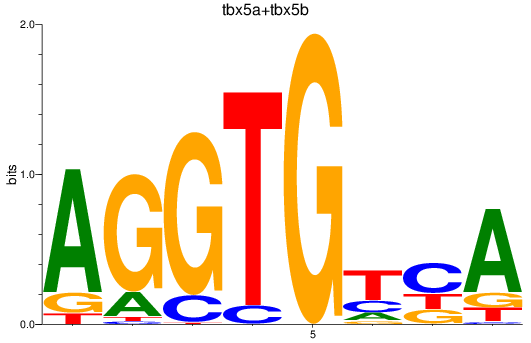
Results for tbx5a+tbx5b
Z-value: 0.91
Transcription factors associated with tbx5a+tbx5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tbx5a
|
ENSDARG00000024894 | T-box transcription factor 5a |
|
tbx5b
|
ENSDARG00000092060 | T-box transcription factor 5b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tbx5b | dr11_v1_chr5_+_23152282_23152282 | -0.18 | 8.5e-02 | Click! |
| tbx5a | dr11_v1_chr5_-_72289648_72289648 | -0.15 | 1.5e-01 | Click! |
Activity profile of tbx5a+tbx5b motif
Sorted Z-values of tbx5a+tbx5b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_32526263 | 14.14 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr21_+_30351256 | 11.44 |
ENSDART00000078341
|
foxi3a
|
forkhead box I3a |
| chr3_+_32526799 | 10.28 |
ENSDART00000185755
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr2_+_2470687 | 10.24 |
ENSDART00000184024
ENSDART00000061955 |
myl13
|
myosin, light chain 13 |
| chr13_-_7031033 | 10.02 |
ENSDART00000193211
|
CABZ01061524.1
|
|
| chr6_-_46941245 | 9.68 |
ENSDART00000037875
|
igfn1.1
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 1 |
| chr7_+_22680560 | 7.26 |
ENSDART00000133761
|
ponzr4
|
plac8 onzin related protein 4 |
| chr18_+_5547185 | 6.86 |
ENSDART00000193977
|
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr21_+_20383837 | 6.80 |
ENSDART00000026430
|
hspb11
|
heat shock protein, alpha-crystallin-related, b11 |
| chr17_-_15611744 | 6.40 |
ENSDART00000010496
|
fhl5
|
four and a half LIM domains 5 |
| chr9_+_23900703 | 6.38 |
ENSDART00000127859
|
trim63b
|
tripartite motif containing 63b |
| chr5_+_57658898 | 5.98 |
ENSDART00000074268
ENSDART00000124568 |
zgc:153929
|
zgc:153929 |
| chr23_-_10254288 | 5.91 |
ENSDART00000081215
|
krt8
|
keratin 8 |
| chr24_-_33703504 | 5.88 |
ENSDART00000079292
|
cavin4b
|
caveolae associated protein 4b |
| chr14_-_17121676 | 5.77 |
ENSDART00000170154
ENSDART00000060479 |
smtnl1
|
smoothelin-like 1 |
| chr10_+_8968203 | 5.63 |
ENSDART00000110443
ENSDART00000080772 |
fstb
|
follistatin b |
| chr15_+_32711663 | 5.49 |
ENSDART00000157854
ENSDART00000167515 |
postnb
|
periostin, osteoblast specific factor b |
| chr11_+_27347076 | 5.47 |
ENSDART00000173383
|
fbln2
|
fibulin 2 |
| chr19_+_19777437 | 5.44 |
ENSDART00000170662
|
hoxa3a
|
homeobox A3a |
| chr8_-_11229523 | 5.42 |
ENSDART00000002164
|
unc45b
|
unc-45 myosin chaperone B |
| chr15_+_32711172 | 5.39 |
ENSDART00000163936
ENSDART00000168135 |
postnb
|
periostin, osteoblast specific factor b |
| chr17_+_2549503 | 5.34 |
ENSDART00000156843
|
si:dkey-248g15.3
|
si:dkey-248g15.3 |
| chr15_-_551177 | 5.26 |
ENSDART00000066774
ENSDART00000154617 |
tagln
|
transgelin |
| chr11_+_29537756 | 5.25 |
ENSDART00000103388
|
wu:fi42e03
|
wu:fi42e03 |
| chr17_+_26965351 | 5.24 |
ENSDART00000114215
ENSDART00000147192 |
grhl3
|
grainyhead-like transcription factor 3 |
| chr23_-_10175898 | 5.02 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr2_+_24304854 | 4.99 |
ENSDART00000078972
|
fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr24_-_10006158 | 4.96 |
ENSDART00000106244
|
zgc:171750
|
zgc:171750 |
| chr5_+_6670945 | 4.91 |
ENSDART00000185686
|
pxna
|
paxillin a |
| chr3_-_12187245 | 4.85 |
ENSDART00000189553
ENSDART00000165131 |
srl
|
sarcalumenin |
| chr1_-_59104145 | 4.73 |
ENSDART00000132495
ENSDART00000152457 |
MFAP4 (1 of many)
si:zfos-2330d3.7
|
si:zfos-2330d3.1 si:zfos-2330d3.7 |
| chr24_-_9997948 | 4.66 |
ENSDART00000136274
|
si:ch211-146l10.7
|
si:ch211-146l10.7 |
| chr24_-_10021341 | 4.66 |
ENSDART00000137250
|
zgc:173856
|
zgc:173856 |
| chr3_+_43086548 | 4.62 |
ENSDART00000163579
|
si:dkey-43p13.5
|
si:dkey-43p13.5 |
| chr23_+_44614056 | 4.61 |
ENSDART00000188379
|
eno3
|
enolase 3, (beta, muscle) |
| chr16_-_31469065 | 4.58 |
ENSDART00000182397
|
si:ch211-251p5.5
|
si:ch211-251p5.5 |
| chr8_+_33035709 | 4.57 |
ENSDART00000131660
|
angptl2b
|
angiopoietin-like 2b |
| chr5_+_26213874 | 4.50 |
ENSDART00000193816
ENSDART00000098514 |
oclnb
|
occludin b |
| chr19_-_34999379 | 4.46 |
ENSDART00000051751
|
zgc:113424
|
zgc:113424 |
| chr21_-_131236 | 4.45 |
ENSDART00000160005
|
si:ch1073-398f15.1
|
si:ch1073-398f15.1 |
| chr24_+_10027902 | 4.43 |
ENSDART00000175961
ENSDART00000172773 |
si:ch211-146l10.8
|
si:ch211-146l10.8 |
| chr5_-_38451082 | 4.37 |
ENSDART00000136428
|
chrne
|
cholinergic receptor, nicotinic, epsilon |
| chr3_+_49397115 | 4.23 |
ENSDART00000176042
|
tecra
|
trans-2,3-enoyl-CoA reductase a |
| chr10_+_19596214 | 4.20 |
ENSDART00000183110
|
CABZ01059626.1
|
|
| chr19_-_38611814 | 4.19 |
ENSDART00000151958
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr9_+_31795343 | 4.17 |
ENSDART00000139584
|
itgbl1
|
integrin, beta-like 1 |
| chr25_+_20077225 | 4.16 |
ENSDART00000136543
|
tnni4b.1
|
troponin I4b, tandem duplicate 1 |
| chr9_-_98982 | 4.15 |
ENSDART00000147882
|
lims2
|
LIM and senescent cell antigen-like domains 2 |
| chr6_+_56141852 | 4.14 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr25_+_8356707 | 4.12 |
ENSDART00000153708
|
muc5.1
|
mucin 5.1, oligomeric mucus/gel-forming |
| chr20_-_26001288 | 4.06 |
ENSDART00000136518
ENSDART00000063177 |
capn3b
|
calpain 3b |
| chr21_+_21374277 | 3.99 |
ENSDART00000079431
|
rtn2b
|
reticulon 2b |
| chr2_+_36007449 | 3.93 |
ENSDART00000161837
|
lamc2
|
laminin, gamma 2 |
| chr23_-_32156278 | 3.89 |
ENSDART00000157479
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr18_+_23373683 | 3.89 |
ENSDART00000001102
ENSDART00000189030 |
mctp2a
|
multiple C2 domains, transmembrane 2a |
| chr24_+_35564668 | 3.86 |
ENSDART00000122734
|
cebpd
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr16_+_3982590 | 3.85 |
ENSDART00000149295
|
zc3h12a
|
zinc finger CCCH-type containing 12A |
| chr20_-_43743700 | 3.83 |
ENSDART00000100620
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr8_+_2478862 | 3.79 |
ENSDART00000131739
|
si:dkeyp-51b9.3
|
si:dkeyp-51b9.3 |
| chr14_+_49251331 | 3.74 |
ENSDART00000148882
|
anxa6
|
annexin A6 |
| chr19_-_5332784 | 3.73 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr19_+_791538 | 3.70 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr8_-_24252933 | 3.69 |
ENSDART00000057624
|
zgc:110353
|
zgc:110353 |
| chr1_+_58205904 | 3.69 |
ENSDART00000147324
|
si:dkey-222h21.12
|
si:dkey-222h21.12 |
| chr7_+_15329819 | 3.66 |
ENSDART00000006018
|
mespaa
|
mesoderm posterior aa |
| chr17_+_24843401 | 3.66 |
ENSDART00000110179
|
cx34.4
|
connexin 34.4 |
| chr21_+_25187210 | 3.64 |
ENSDART00000101147
ENSDART00000167528 |
si:dkey-183i3.5
|
si:dkey-183i3.5 |
| chr19_-_2861444 | 3.62 |
ENSDART00000169053
|
clec3bb
|
C-type lectin domain family 3, member Bb |
| chr7_-_2047639 | 3.61 |
ENSDART00000173892
|
si:cabz01007794.1
|
si:cabz01007794.1 |
| chr12_-_46228023 | 3.61 |
ENSDART00000153455
|
si:ch211-226h7.6
|
si:ch211-226h7.6 |
| chr10_+_19595009 | 3.50 |
ENSDART00000112276
|
zgc:173837
|
zgc:173837 |
| chr17_-_6349044 | 3.45 |
ENSDART00000081707
|
oct2
|
organic cation transporter 2 |
| chr13_-_37180815 | 3.39 |
ENSDART00000139907
|
si:dkeyp-77c8.1
|
si:dkeyp-77c8.1 |
| chr9_+_17306162 | 3.34 |
ENSDART00000075926
|
scel
|
sciellin |
| chr1_+_40308077 | 3.34 |
ENSDART00000138992
|
vwa10.2
|
von Willebrand factor A domain containing 10, tandem duplicate 2 |
| chr16_+_26774182 | 3.28 |
ENSDART00000042895
|
cdh17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr3_-_39171968 | 3.26 |
ENSDART00000154494
|
si:dkeyp-57f11.2
|
si:dkeyp-57f11.2 |
| chr18_+_54354 | 3.25 |
ENSDART00000097163
|
zgc:158482
|
zgc:158482 |
| chr5_-_64355227 | 3.18 |
ENSDART00000170787
|
fam78aa
|
family with sequence similarity 78, member Aa |
| chr9_-_30165621 | 3.18 |
ENSDART00000089543
|
abi3bpa
|
ABI family, member 3 (NESH) binding protein a |
| chr14_+_48862987 | 3.14 |
ENSDART00000167810
|
zgc:154054
|
zgc:154054 |
| chr21_-_34972872 | 3.03 |
ENSDART00000023838
|
lipia
|
lipase, member Ia |
| chr7_-_69795488 | 3.02 |
ENSDART00000162414
|
USP53 (1 of many)
|
ubiquitin specific peptidase 53 |
| chr12_+_20641102 | 3.00 |
ENSDART00000152964
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr23_-_5719453 | 2.99 |
ENSDART00000033093
|
lad1
|
ladinin |
| chr8_-_36412936 | 2.95 |
ENSDART00000159276
|
si:zfos-2070c2.3
|
si:zfos-2070c2.3 |
| chr1_+_24355515 | 2.93 |
ENSDART00000051763
|
rps3a
|
ribosomal protein S3A |
| chr10_-_13116337 | 2.92 |
ENSDART00000164568
|
musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr8_-_3312384 | 2.88 |
ENSDART00000035965
|
fut9b
|
fucosyltransferase 9b |
| chr21_+_33454147 | 2.86 |
ENSDART00000053208
|
rps14
|
ribosomal protein S14 |
| chr19_+_5640504 | 2.84 |
ENSDART00000179987
|
ft2
|
alpha(1,3)fucosyltransferase gene 2 |
| chr25_+_31405266 | 2.81 |
ENSDART00000103395
|
tnnt3a
|
troponin T type 3a (skeletal, fast) |
| chr22_+_26600834 | 2.80 |
ENSDART00000157411
|
adcy9
|
adenylate cyclase 9 |
| chr21_-_11632403 | 2.78 |
ENSDART00000171708
ENSDART00000138619 ENSDART00000136308 ENSDART00000144770 |
cast
|
calpastatin |
| chr15_+_36054864 | 2.74 |
ENSDART00000156697
|
col4a3
|
collagen, type IV, alpha 3 |
| chr4_-_77114795 | 2.70 |
ENSDART00000144849
|
CU467646.2
|
|
| chr11_-_25853212 | 2.68 |
ENSDART00000145655
|
tmem51b
|
transmembrane protein 51b |
| chr13_-_34858500 | 2.67 |
ENSDART00000184843
|
sptlc3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr2_-_28396993 | 2.65 |
ENSDART00000188170
|
CABZ01056052.1
|
|
| chr12_+_28749189 | 2.63 |
ENSDART00000013980
|
tbx21
|
T-box 21 |
| chr19_+_34952582 | 2.63 |
ENSDART00000147714
|
wisp1a
|
WNT1 inducible signaling pathway protein 1a |
| chr3_+_36275633 | 2.61 |
ENSDART00000185027
ENSDART00000149532 ENSDART00000102883 ENSDART00000148444 |
zgc:86896
|
zgc:86896 |
| chr5_-_16475374 | 2.58 |
ENSDART00000134274
ENSDART00000136004 |
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr4_-_11594202 | 2.58 |
ENSDART00000002682
|
net1
|
neuroepithelial cell transforming 1 |
| chr6_+_23057311 | 2.56 |
ENSDART00000026448
|
evpla
|
envoplakin a |
| chr16_-_30878521 | 2.55 |
ENSDART00000141403
|
dennd3b
|
DENN/MADD domain containing 3b |
| chr10_+_19569052 | 2.55 |
ENSDART00000058425
|
CABZ01059627.1
|
|
| chr1_+_54655160 | 2.48 |
ENSDART00000190319
|
si:ch211-202h22.7
|
si:ch211-202h22.7 |
| chr3_-_32603191 | 2.46 |
ENSDART00000150997
|
si:ch73-248e21.7
|
si:ch73-248e21.7 |
| chr3_+_5331428 | 2.45 |
ENSDART00000156561
|
si:ch73-106l15.4
|
si:ch73-106l15.4 |
| chr4_-_64703 | 2.45 |
ENSDART00000167851
|
CU856344.1
|
|
| chr3_-_53508580 | 2.42 |
ENSDART00000073978
|
zgc:171711
|
zgc:171711 |
| chr13_+_18533005 | 2.40 |
ENSDART00000136024
|
ftr14l
|
finTRIM family, member 14-like |
| chr3_-_60027255 | 2.38 |
ENSDART00000189252
ENSDART00000154684 |
recql5
|
RecQ helicase-like 5 |
| chr15_+_13984879 | 2.36 |
ENSDART00000159438
|
zgc:162730
|
zgc:162730 |
| chr14_-_28568107 | 2.36 |
ENSDART00000042850
ENSDART00000145502 |
insb
|
preproinsulin b |
| chr6_+_40794015 | 2.34 |
ENSDART00000144479
|
gata2b
|
GATA binding protein 2b |
| chr12_-_36045283 | 2.34 |
ENSDART00000160646
|
gprc5c
|
G protein-coupled receptor, class C, group 5, member C |
| chr19_-_34995629 | 2.34 |
ENSDART00000141704
|
si:rp71-45k5.2
|
si:rp71-45k5.2 |
| chr15_-_36727462 | 2.31 |
ENSDART00000085971
|
nphs1
|
nephrosis 1, congenital, Finnish type (nephrin) |
| chr23_-_29003864 | 2.30 |
ENSDART00000148257
|
casz1
|
castor zinc finger 1 |
| chr14_+_21934331 | 2.28 |
ENSDART00000123225
|
zgc:113229
|
zgc:113229 |
| chr11_+_21050326 | 2.24 |
ENSDART00000065984
|
zgc:113307
|
zgc:113307 |
| chr17_-_53359028 | 2.24 |
ENSDART00000185218
|
CABZ01068208.1
|
|
| chr18_-_14274803 | 2.22 |
ENSDART00000166643
|
mlycd
|
malonyl-CoA decarboxylase |
| chr4_+_14981854 | 2.22 |
ENSDART00000067046
|
cax1
|
cation/H+ exchanger protein 1 |
| chr19_+_4076259 | 2.21 |
ENSDART00000160633
|
zgc:173581
|
zgc:173581 |
| chr21_-_41025340 | 2.20 |
ENSDART00000148231
|
plac8l1
|
PLAC8-like 1 |
| chr5_-_32338866 | 2.17 |
ENSDART00000017956
ENSDART00000047670 |
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr3_+_5297493 | 2.17 |
ENSDART00000138596
|
si:ch211-150d5.3
|
si:ch211-150d5.3 |
| chr3_-_5228841 | 2.16 |
ENSDART00000092373
ENSDART00000182438 |
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr20_+_52546186 | 2.14 |
ENSDART00000110777
ENSDART00000153377 ENSDART00000153013 ENSDART00000042704 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr25_-_35169303 | 2.13 |
ENSDART00000193240
|
ano9a
|
anoctamin 9a |
| chr23_+_19790962 | 2.12 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr24_-_42108655 | 2.12 |
ENSDART00000032722
|
pigm
|
phosphatidylinositol glycan anchor biosynthesis, class M |
| chr7_-_58776400 | 2.11 |
ENSDART00000167433
|
sox17
|
SRY (sex determining region Y)-box 17 |
| chr2_-_2642476 | 2.11 |
ENSDART00000124032
|
serbp1b
|
SERPINE1 mRNA binding protein 1b |
| chr10_-_21542702 | 2.09 |
ENSDART00000146761
ENSDART00000134502 |
zgc:165539
|
zgc:165539 |
| chr12_-_44180132 | 2.09 |
ENSDART00000165998
|
si:ch73-329n5.1
|
si:ch73-329n5.1 |
| chr23_-_270847 | 2.08 |
ENSDART00000191867
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr23_-_16734009 | 2.07 |
ENSDART00000125449
|
si:ch211-224l10.4
|
si:ch211-224l10.4 |
| chr7_+_65673885 | 2.06 |
ENSDART00000169182
|
parvab
|
parvin, alpha b |
| chr10_-_1961930 | 2.06 |
ENSDART00000122446
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr3_-_23604396 | 2.05 |
ENSDART00000078423
|
atp5mc1
|
ATP synthase membrane subunit c locus 1 |
| chr3_+_26345732 | 2.03 |
ENSDART00000128613
|
rps15a
|
ribosomal protein S15a |
| chr4_-_880415 | 2.03 |
ENSDART00000149162
|
aim1b
|
crystallin beta-gamma domain containing 1b |
| chr22_-_29242347 | 2.02 |
ENSDART00000040761
|
pvalb7
|
parvalbumin 7 |
| chr23_-_45501177 | 2.00 |
ENSDART00000150103
|
col24a1
|
collagen type XXIV alpha 1 |
| chr5_-_68333081 | 1.98 |
ENSDART00000168786
|
h1m
|
linker histone H1M |
| chr3_+_33761549 | 1.95 |
ENSDART00000169337
|
ier2a
|
immediate early response 2a |
| chr2_+_30379650 | 1.95 |
ENSDART00000129542
|
crispld1b
|
cysteine-rich secretory protein LCCL domain containing 1b |
| chr15_-_20916251 | 1.94 |
ENSDART00000134053
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr8_-_20243389 | 1.94 |
ENSDART00000184904
|
acer1
|
alkaline ceramidase 1 |
| chr11_-_11575070 | 1.92 |
ENSDART00000142208
|
zgc:110712
|
zgc:110712 |
| chr23_+_31107685 | 1.89 |
ENSDART00000103448
|
tbx18
|
T-box 18 |
| chr13_+_23623346 | 1.88 |
ENSDART00000057619
|
il22ra2
|
interleukin 22 receptor, alpha 2 |
| chr7_-_55648336 | 1.88 |
ENSDART00000147792
ENSDART00000135304 ENSDART00000131923 |
pabpn1l
|
poly(A) binding protein, nuclear 1-like (cytoplasmic) |
| chr14_+_7699443 | 1.85 |
ENSDART00000123139
|
brd8
|
bromodomain containing 8 |
| chr13_-_51846224 | 1.85 |
ENSDART00000184663
|
LT631684.2
|
|
| chr10_-_15849027 | 1.83 |
ENSDART00000184682
|
tjp2a
|
tight junction protein 2a (zona occludens 2) |
| chr5_+_36439405 | 1.83 |
ENSDART00000102973
|
eda
|
ectodysplasin A |
| chr19_-_3741602 | 1.83 |
ENSDART00000170301
|
btr22
|
bloodthirsty-related gene family, member 22 |
| chr7_-_51368681 | 1.83 |
ENSDART00000146385
|
arhgap36
|
Rho GTPase activating protein 36 |
| chr14_+_41318881 | 1.82 |
ENSDART00000192137
|
xkrx
|
XK, Kell blood group complex subunit-related, X-linked |
| chr5_+_51909740 | 1.82 |
ENSDART00000162541
|
thbs4a
|
thrombospondin 4a |
| chr23_-_26535875 | 1.80 |
ENSDART00000135988
|
si:dkey-205h13.2
|
si:dkey-205h13.2 |
| chr15_-_28587490 | 1.80 |
ENSDART00000186196
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr10_-_35149513 | 1.80 |
ENSDART00000063434
ENSDART00000131291 |
ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr16_+_24971717 | 1.79 |
ENSDART00000156958
|
fpr1
|
formyl peptide receptor 1 |
| chr2_-_32237916 | 1.78 |
ENSDART00000141418
|
fam49ba
|
family with sequence similarity 49, member Ba |
| chr8_+_40628926 | 1.77 |
ENSDART00000163598
|
dusp2
|
dual specificity phosphatase 2 |
| chr11_+_36683859 | 1.75 |
ENSDART00000170102
|
si:ch211-11c3.12
|
si:ch211-11c3.12 |
| chr14_-_48103207 | 1.74 |
ENSDART00000056712
|
etfdh
|
electron-transferring-flavoprotein dehydrogenase |
| chr5_-_16475682 | 1.74 |
ENSDART00000090695
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr11_-_6452444 | 1.73 |
ENSDART00000137879
ENSDART00000134957 ENSDART00000004483 |
larp6b
|
La ribonucleoprotein domain family, member 6b |
| chr8_+_25569903 | 1.72 |
ENSDART00000062375
|
si:dkey-48j7.3
|
si:dkey-48j7.3 |
| chr4_+_8638622 | 1.71 |
ENSDART00000186829
|
wnt5b
|
wingless-type MMTV integration site family, member 5b |
| chr1_-_6085750 | 1.70 |
ENSDART00000138891
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr13_+_27232694 | 1.68 |
ENSDART00000131128
|
rin2
|
Ras and Rab interactor 2 |
| chr2_-_14793343 | 1.67 |
ENSDART00000132264
|
si:ch73-366i20.1
|
si:ch73-366i20.1 |
| chr2_-_58257624 | 1.66 |
ENSDART00000098940
|
foxl2b
|
forkhead box L2b |
| chr4_+_8532580 | 1.66 |
ENSDART00000162065
|
wnt5b
|
wingless-type MMTV integration site family, member 5b |
| chr14_+_35024521 | 1.65 |
ENSDART00000158634
ENSDART00000170631 |
ebf3a
|
early B cell factor 3a |
| chr22_-_17652914 | 1.65 |
ENSDART00000138483
|
si:ch73-243b8.4
|
si:ch73-243b8.4 |
| chr22_-_17653143 | 1.64 |
ENSDART00000089171
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr24_-_2423791 | 1.63 |
ENSDART00000190402
|
rreb1a
|
ras responsive element binding protein 1a |
| chr10_-_45029041 | 1.63 |
ENSDART00000167878
|
polm
|
polymerase (DNA directed), mu |
| chr20_+_51104367 | 1.63 |
ENSDART00000073981
|
eif2s1b
|
eukaryotic translation initiation factor 2, subunit 1 alpha b |
| chr23_-_20345473 | 1.63 |
ENSDART00000140935
|
si:rp71-17i16.6
|
si:rp71-17i16.6 |
| chr3_+_12748038 | 1.63 |
ENSDART00000181174
|
cyp2k21
|
cytochrome P450, family 2, subfamily k, polypeptide 21 |
| chr25_+_18556588 | 1.62 |
ENSDART00000073726
|
cav2
|
caveolin 2 |
| chr1_+_524717 | 1.61 |
ENSDART00000102421
ENSDART00000184473 |
mrpl16
|
mitochondrial ribosomal protein L16 |
| chr7_+_41887429 | 1.61 |
ENSDART00000115090
|
gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr23_+_23232136 | 1.60 |
ENSDART00000126479
ENSDART00000187764 |
plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr3_-_12352793 | 1.60 |
ENSDART00000165013
|
pam16
|
presequence translocase associated motor 16 |
| chr9_-_15362556 | 1.59 |
ENSDART00000191233
|
fn1a
|
fibronectin 1a |
| chr16_+_5926520 | 1.59 |
ENSDART00000162229
|
ulk4
|
unc-51 like kinase 4 |
| chr21_-_3700334 | 1.59 |
ENSDART00000137844
|
atp8b1
|
ATPase phospholipid transporting 8B1 |
| chr19_+_41464870 | 1.59 |
ENSDART00000102778
|
dlx6a
|
distal-less homeobox 6a |
| chr4_-_77216726 | 1.59 |
ENSDART00000099943
|
psmb10
|
proteasome subunit beta 10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of tbx5a+tbx5b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 11.4 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 1.7 | 6.9 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 1.4 | 5.5 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 1.4 | 4.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 1.1 | 4.3 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 1.0 | 6.2 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 1.0 | 5.9 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.9 | 3.7 | GO:0003418 | plasma membrane repair(GO:0001778) chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.8 | 5.4 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.7 | 2.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.7 | 2.1 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.7 | 3.4 | GO:1904105 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.7 | 10.7 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.6 | 3.8 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.6 | 1.7 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.5 | 4.1 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.5 | 1.6 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.5 | 5.6 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.5 | 3.7 | GO:0031179 | peptide modification(GO:0031179) |
| 0.4 | 2.2 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.4 | 3.9 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.4 | 4.6 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.4 | 2.8 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.4 | 1.2 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.4 | 1.5 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.4 | 2.9 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.4 | 2.2 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.4 | 1.4 | GO:0006266 | DNA ligation(GO:0006266) DNA ligation involved in DNA repair(GO:0051103) |
| 0.4 | 1.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.3 | 1.4 | GO:0070199 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.3 | 2.4 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.3 | 4.9 | GO:0061055 | myotome development(GO:0061055) |
| 0.3 | 1.9 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.3 | 4.1 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.3 | 6.8 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.3 | 1.8 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) |
| 0.3 | 4.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.3 | 4.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.3 | 0.8 | GO:0098581 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.3 | 2.2 | GO:1905207 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.3 | 1.3 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.3 | 1.3 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.3 | 1.6 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.3 | 1.5 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.2 | 1.2 | GO:0006031 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.2 | 1.6 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.2 | 1.6 | GO:0006953 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) acute-phase response(GO:0006953) establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.2 | 1.1 | GO:0035678 | neuromast hair cell morphogenesis(GO:0035678) |
| 0.2 | 4.4 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.2 | 1.3 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.2 | 2.6 | GO:0032925 | regulation of activin receptor signaling pathway(GO:0032925) |
| 0.2 | 0.6 | GO:0098725 | somatic stem cell population maintenance(GO:0035019) symmetric cell division(GO:0098725) |
| 0.2 | 2.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.2 | 0.6 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.2 | 0.6 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.2 | 0.6 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.2 | 0.8 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 | 1.0 | GO:0072672 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.2 | 0.7 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.2 | 0.5 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.2 | 3.0 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.2 | 1.9 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.2 | 5.2 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.2 | 0.9 | GO:0034695 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.2 | 0.5 | GO:0030238 | female sex determination(GO:0030237) male sex determination(GO:0030238) |
| 0.2 | 0.7 | GO:2000392 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.2 | 2.6 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.2 | 1.0 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.2 | 0.7 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.2 | 3.0 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.2 | 1.1 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.2 | 0.9 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.2 | 1.4 | GO:0060004 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.2 | 0.6 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.2 | 1.8 | GO:0051121 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 1.0 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.1 | 2.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.0 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.1 | 6.4 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.1 | 0.7 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 2.5 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 1.3 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 2.1 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 0.4 | GO:0001774 | microglial cell activation(GO:0001774) |
| 0.1 | 4.2 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 2.3 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 1.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 2.9 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 1.1 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 2.8 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.1 | 2.1 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.1 | 2.0 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 4.2 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 2.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 3.9 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.1 | 0.6 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.5 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 1.6 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.4 | GO:0055024 | regulation of cardiac muscle tissue development(GO:0055024) |
| 0.1 | 0.9 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.1 | 1.5 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.1 | 1.0 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 2.1 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 2.8 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 1.1 | GO:0010092 | specification of organ identity(GO:0010092) |
| 0.1 | 2.0 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.1 | 1.8 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.6 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 1.2 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 2.0 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 1.8 | GO:0034112 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.1 | 2.4 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 3.9 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 2.6 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.1 | 3.6 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.1 | 2.1 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 0.8 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 1.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 1.4 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 0.5 | GO:0036372 | opsin transport(GO:0036372) |
| 0.1 | 0.8 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 0.3 | GO:0042148 | strand invasion(GO:0042148) |
| 0.1 | 0.3 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.6 | GO:0042664 | negative regulation of endodermal cell fate specification(GO:0042664) negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 1.2 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.1 | 0.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.2 | GO:0050881 | multicellular organismal movement(GO:0050879) musculoskeletal movement(GO:0050881) |
| 0.1 | 1.6 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.1 | 2.6 | GO:0045104 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.1 | 0.5 | GO:0070874 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 1.4 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.1 | 1.1 | GO:0008406 | gonad development(GO:0008406) regulation of meiotic cell cycle(GO:0051445) |
| 0.1 | 1.2 | GO:0050935 | iridophore differentiation(GO:0050935) |
| 0.1 | 0.5 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 1.4 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 0.2 | GO:0043011 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 1.7 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 0.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.3 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.1 | 0.6 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 11.6 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.1 | 1.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 1.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.3 | GO:2000463 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.9 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 4.0 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.5 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.2 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.3 | GO:0010595 | positive regulation of endothelial cell migration(GO:0010595) |
| 0.0 | 1.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.2 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 1.0 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.8 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.3 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.2 | GO:0061157 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.0 | 0.5 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 0.9 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 1.8 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.3 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 1.5 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.1 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 2.3 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 1.5 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.9 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.4 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 2.6 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 0.2 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 3.8 | GO:0045055 | regulated exocytosis(GO:0045055) |
| 0.0 | 0.6 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.5 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.7 | GO:0019229 | regulation of vasoconstriction(GO:0019229) |
| 0.0 | 0.1 | GO:0070197 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.2 | GO:0002285 | lymphocyte activation involved in immune response(GO:0002285) T cell activation involved in immune response(GO:0002286) positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.4 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.1 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.7 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.9 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 2.0 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 2.3 | GO:0001708 | cell fate specification(GO:0001708) |
| 0.0 | 1.1 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 1.8 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 4.4 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.4 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.8 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.4 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 2.5 | GO:0048793 | pronephros development(GO:0048793) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.3 | GO:1902307 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.9 | GO:0043648 | dicarboxylic acid metabolic process(GO:0043648) |
| 0.0 | 1.3 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 1.5 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.5 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.3 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 0.3 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 1.3 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.1 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 1.2 | GO:0008544 | epidermis development(GO:0008544) |
| 0.0 | 2.9 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.0 | 0.2 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.2 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.3 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 7.4 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.3 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 10.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.9 | 4.3 | GO:1990923 | PET complex(GO:1990923) |
| 0.6 | 1.9 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.5 | 1.6 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.5 | 1.4 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.4 | 2.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.4 | 2.3 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.4 | 2.2 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.4 | 4.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.3 | 1.4 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.3 | 0.9 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.3 | 2.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.3 | 0.9 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.3 | 1.1 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.3 | 3.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.3 | 0.8 | GO:0000941 | condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 0.3 | 2.7 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.2 | 4.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 1.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 2.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.2 | 5.4 | GO:0031672 | A band(GO:0031672) |
| 0.2 | 7.5 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.2 | 8.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 0.7 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.2 | 2.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 7.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 20.8 | GO:0030055 | cell-substrate junction(GO:0030055) |
| 0.2 | 7.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.2 | GO:0030428 | cell septum(GO:0030428) |
| 0.1 | 11.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.5 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 2.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 1.4 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 7.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 0.3 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 0.9 | GO:0045240 | dihydrolipoyl dehydrogenase complex(GO:0045240) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 1.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.6 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.9 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 1.0 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 26.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 0.2 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 3.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.5 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.9 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 1.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 0.4 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 2.1 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 2.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.1 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 4.7 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.7 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 3.4 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 2.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.5 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 1.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.9 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 20.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 1.3 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 6.9 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.6 | 1.7 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.6 | 2.2 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.5 | 4.3 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.5 | 1.6 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.5 | 1.6 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.4 | 2.6 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.4 | 3.7 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.4 | 2.9 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.4 | 6.0 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.4 | 2.8 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.4 | 2.7 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.4 | 1.4 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.4 | 3.9 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.3 | 3.0 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.3 | 1.9 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.3 | 5.6 | GO:0048185 | activin binding(GO:0048185) |
| 0.3 | 0.8 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.3 | 3.0 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.3 | 0.8 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.3 | 2.1 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.3 | 2.4 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.3 | 1.3 | GO:0015140 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.3 | 2.8 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.2 | 0.7 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.2 | 1.2 | GO:0098639 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.2 | 2.8 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.2 | 1.6 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.2 | 3.3 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.2 | 4.3 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.2 | 3.0 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.2 | 1.8 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 1.6 | GO:0001948 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.2 | 5.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 1.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 0.5 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.2 | 0.9 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 1.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.2 | 0.7 | GO:0004945 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.2 | 1.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.2 | 4.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.2 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.1 | 1.9 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.1 | 1.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 1.1 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 4.2 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 1.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.8 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 4.4 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.1 | 1.4 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 0.9 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 0.9 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 2.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 2.3 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 1.0 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 0.6 | GO:0035620 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.1 | 0.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.9 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 2.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.9 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 4.9 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.1 | 2.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 1.0 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 1.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.5 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.1 | 0.7 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 10.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 3.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.6 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 4.2 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 14.8 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.1 | 1.9 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 0.3 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 4.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 2.8 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.9 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 1.0 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 0.2 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.1 | 0.6 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 1.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 4.2 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.1 | 0.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.6 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 1.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.5 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 1.3 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.5 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 1.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 1.4 | GO:0099604 | ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 1.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 1.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 1.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 5.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 3.3 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 5.8 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.6 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.6 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 1.1 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 2.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 2.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.5 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.4 | GO:0005223 | intracellular ligand-gated ion channel activity(GO:0005217) intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 3.3 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 4.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 4.1 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 0.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 8.6 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.4 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.4 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.6 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 8.8 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 34.3 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 2.3 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 1.5 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 1.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 8.7 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) phosphoprotein binding(GO:0051219) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.3 | 3.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.3 | 0.6 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.2 | 8.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 1.0 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.2 | 3.9 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 2.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 7.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 3.2 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 3.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.6 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 2.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 3.3 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 2.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 2.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 1.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 0.3 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 1.3 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 0.9 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 1.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 1.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 0.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 1.5 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.1 | 2.3 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 1.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 2.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 6.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 6.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 2.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.2 | PID SHP2 PATHWAY | SHP2 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.3 | 2.7 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 1.4 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.2 | 0.6 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 2.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 2.8 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.2 | 1.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 8.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 8.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.2 | 1.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.7 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 2.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 1.3 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 3.0 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 0.2 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.1 | 0.6 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 0.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 3.9 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 0.8 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 0.9 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 3.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 0.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.0 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.5 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.1 | 0.5 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 3.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 1.0 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.9 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 2.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 2.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.3 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |