Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
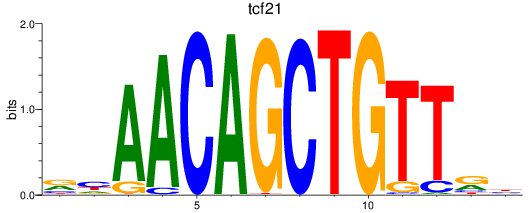
Results for tcf21
Z-value: 1.11
Transcription factors associated with tcf21
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tcf21
|
ENSDARG00000036869 | transcription factor 21 |
|
tcf21
|
ENSDARG00000111209 | transcription factor 21 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tcf21 | dr11_v1_chr23_-_31555696_31555696 | 0.09 | 3.8e-01 | Click! |
Activity profile of tcf21 motif
Sorted Z-values of tcf21 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_28830643 | 32.01 |
ENSDART00000051448
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr5_+_28849155 | 27.67 |
ENSDART00000079090
|
zgc:174259
|
zgc:174259 |
| chr5_+_28858345 | 22.22 |
ENSDART00000111180
|
si:ch211-186e20.2
|
si:ch211-186e20.2 |
| chr11_-_5865744 | 19.16 |
ENSDART00000104360
|
gamt
|
guanidinoacetate N-methyltransferase |
| chr3_+_1055588 | 19.15 |
ENSDART00000143220
|
zgc:153921
|
zgc:153921 |
| chr5_+_28848870 | 14.22 |
ENSDART00000149563
|
zgc:174259
|
zgc:174259 |
| chr5_+_28857969 | 13.66 |
ENSDART00000149850
|
si:ch211-186e20.2
|
si:ch211-186e20.2 |
| chr7_+_6652967 | 13.49 |
ENSDART00000102681
|
pnp5a
|
purine nucleoside phosphorylase 5a |
| chr16_+_23972126 | 13.27 |
ENSDART00000132742
ENSDART00000145330 |
apoc1
|
apolipoprotein C-I |
| chr5_+_28830388 | 13.06 |
ENSDART00000149150
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr7_+_34794829 | 12.75 |
ENSDART00000009698
ENSDART00000075089 ENSDART00000173456 |
esrp2
|
epithelial splicing regulatory protein 2 |
| chr8_+_39724138 | 12.63 |
ENSDART00000009323
|
pla2g1b
|
phospholipase A2, group IB (pancreas) |
| chr6_+_112579 | 11.76 |
ENSDART00000034505
|
ap1m2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr7_+_10610791 | 11.39 |
ENSDART00000166064
|
fah
|
fumarylacetoacetate hydrolase (fumarylacetoacetase) |
| chr20_+_42668875 | 10.43 |
ENSDART00000048890
|
slc22a2
|
solute carrier family 22 (organic cation transporter), member 2 |
| chr3_-_21094437 | 8.67 |
ENSDART00000153739
ENSDART00000109790 |
nlk1
|
nemo-like kinase, type 1 |
| chr2_+_4208323 | 7.68 |
ENSDART00000167906
|
gata6
|
GATA binding protein 6 |
| chr7_-_24364536 | 7.67 |
ENSDART00000064789
|
txn
|
thioredoxin |
| chr9_-_35633827 | 7.15 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr8_-_27687095 | 6.20 |
ENSDART00000086946
|
mov10b.1
|
Moloney leukemia virus 10b, tandem duplicate 1 |
| chr8_+_24747865 | 6.15 |
ENSDART00000078656
|
slc16a4
|
solute carrier family 16, member 4 |
| chr7_+_49664174 | 6.13 |
ENSDART00000137059
ENSDART00000131210 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr25_+_3326885 | 6.06 |
ENSDART00000104866
|
ldhbb
|
lactate dehydrogenase Bb |
| chr15_+_20403084 | 6.04 |
ENSDART00000141388
ENSDART00000152734 |
cox7a1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr24_+_22056386 | 5.97 |
ENSDART00000132390
|
ankrd33ba
|
ankyrin repeat domain 33ba |
| chr9_+_21146862 | 5.90 |
ENSDART00000136365
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr5_+_67448664 | 5.88 |
ENSDART00000105731
|
si:dkey-251i10.1
|
si:dkey-251i10.1 |
| chr5_-_29152457 | 5.72 |
ENSDART00000078469
|
noxa1
|
NADPH oxidase activator 1 |
| chr24_+_37709191 | 5.63 |
ENSDART00000066558
|
decr2
|
2,4-dienoyl CoA reductase 2, peroxisomal |
| chr19_+_43119014 | 5.59 |
ENSDART00000023156
|
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr11_+_44503774 | 5.55 |
ENSDART00000169295
|
ero1b
|
endoplasmic reticulum oxidoreductase beta |
| chr7_+_44715224 | 5.39 |
ENSDART00000184630
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr6_-_54348568 | 5.08 |
ENSDART00000156501
|
zgc:101577
|
zgc:101577 |
| chr15_-_39969988 | 5.01 |
ENSDART00000146054
|
rps5
|
ribosomal protein S5 |
| chr1_-_52498146 | 4.95 |
ENSDART00000122217
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr16_-_31686602 | 4.93 |
ENSDART00000170357
|
c1s
|
complement component 1, s subcomponent |
| chr12_-_33859950 | 4.60 |
ENSDART00000131181
|
sema4gb
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Gb |
| chr1_-_52497834 | 4.54 |
ENSDART00000136469
ENSDART00000004233 |
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr10_-_25217347 | 4.53 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr8_-_19280856 | 4.50 |
ENSDART00000100473
|
zgc:77486
|
zgc:77486 |
| chr10_+_31809226 | 4.43 |
ENSDART00000087898
|
foxo1b
|
forkhead box O1 b |
| chr4_-_75175407 | 4.33 |
ENSDART00000180125
|
CABZ01043953.1
|
|
| chr21_-_20929575 | 4.30 |
ENSDART00000163889
|
c6
|
complement component 6 |
| chr19_+_7636941 | 4.29 |
ENSDART00000081611
ENSDART00000163805 ENSDART00000112404 |
cgnb
|
cingulin b |
| chr5_+_25762271 | 4.27 |
ENSDART00000181323
|
tmem2
|
transmembrane protein 2 |
| chr25_-_7650335 | 4.24 |
ENSDART00000089034
|
myo5ab
|
myosin VAb |
| chr2_-_17114852 | 4.23 |
ENSDART00000006549
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr11_+_21910343 | 4.18 |
ENSDART00000161485
|
foxp4
|
forkhead box P4 |
| chr1_+_52560549 | 4.18 |
ENSDART00000167514
|
abca1a
|
ATP-binding cassette, sub-family A (ABC1), member 1A |
| chr15_+_29276508 | 3.99 |
ENSDART00000170537
ENSDART00000126559 |
rap1gap2a
|
RAP1 GTPase activating protein 2a |
| chr8_+_45334255 | 3.82 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr14_-_34605607 | 3.81 |
ENSDART00000191608
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr22_-_16154771 | 3.76 |
ENSDART00000009464
|
slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr13_+_40770628 | 3.45 |
ENSDART00000085846
|
nkx1.2la
|
NK1 transcription factor related 2-like,a |
| chr21_-_37435162 | 3.43 |
ENSDART00000133585
|
fam114a2
|
family with sequence similarity 114, member A2 |
| chr19_+_43119698 | 3.36 |
ENSDART00000167847
ENSDART00000186962 ENSDART00000187305 |
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr20_-_26551210 | 3.36 |
ENSDART00000077715
|
si:dkey-25e12.3
|
si:dkey-25e12.3 |
| chr6_+_42475730 | 3.34 |
ENSDART00000150226
|
mst1ra
|
macrophage stimulating 1 receptor a |
| chr2_-_17115256 | 3.25 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr15_-_18067220 | 3.23 |
ENSDART00000113142
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr2_+_32016516 | 3.22 |
ENSDART00000135040
|
mycb
|
MYC proto-oncogene, bHLH transcription factor b |
| chr10_-_44560165 | 3.13 |
ENSDART00000181217
ENSDART00000076084 |
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr21_-_293146 | 3.12 |
ENSDART00000157781
|
plgrkt
|
plasminogen receptor, C-terminal lysine transmembrane protein |
| chr15_-_25365319 | 3.10 |
ENSDART00000152651
|
cluha
|
clustered mitochondria (cluA/CLU1) homolog a |
| chr21_+_19418563 | 3.09 |
ENSDART00000181113
ENSDART00000080110 |
amacr
|
alpha-methylacyl-CoA racemase |
| chr1_+_33390286 | 3.05 |
ENSDART00000139004
|
dhrsx
|
dehydrogenase/reductase (SDR family) X-linked |
| chr2_+_32016256 | 2.96 |
ENSDART00000005143
|
mycb
|
MYC proto-oncogene, bHLH transcription factor b |
| chr7_-_17690756 | 2.92 |
ENSDART00000173759
|
map4k2
|
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr10_+_29849977 | 2.91 |
ENSDART00000180242
|
hspa8
|
heat shock protein 8 |
| chr17_+_8323348 | 2.90 |
ENSDART00000169900
|
cdc42bpaa
|
CDC42 binding protein kinase alpha (DMPK-like) a |
| chr7_-_24046999 | 2.88 |
ENSDART00000144616
ENSDART00000124653 ENSDART00000127813 |
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr13_+_11876437 | 2.87 |
ENSDART00000179753
|
trim8a
|
tripartite motif containing 8a |
| chr9_-_12811936 | 2.87 |
ENSDART00000188490
|
myo10l3
|
myosin X-like 3 |
| chr14_-_34605804 | 2.86 |
ENSDART00000144547
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr3_-_14571514 | 2.85 |
ENSDART00000137197
|
swsap1
|
SWIM-type zinc finger 7 associated protein 1 |
| chr5_-_13315726 | 2.83 |
ENSDART00000143364
|
sdf2l1
|
stromal cell-derived factor 2-like 1 |
| chr3_-_34724879 | 2.82 |
ENSDART00000177021
|
thraa
|
thyroid hormone receptor alpha a |
| chr10_+_29850330 | 2.75 |
ENSDART00000168898
|
hspa8
|
heat shock protein 8 |
| chr24_-_25166416 | 2.74 |
ENSDART00000111552
ENSDART00000169495 |
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr21_-_45685063 | 2.67 |
ENSDART00000162422
|
sar1b
|
secretion associated, Ras related GTPase 1B |
| chr17_+_53156530 | 2.66 |
ENSDART00000126277
ENSDART00000156774 |
dph6
|
diphthamine biosynthesis 6 |
| chr13_+_24584401 | 2.63 |
ENSDART00000057599
|
fuom
|
fucose mutarotase |
| chr9_-_49531762 | 2.61 |
ENSDART00000121875
|
xirp2b
|
xin actin binding repeat containing 2b |
| chr15_+_6459847 | 2.57 |
ENSDART00000157250
ENSDART00000065824 |
bace2
|
beta-site APP-cleaving enzyme 2 |
| chr18_-_7948188 | 2.55 |
ENSDART00000091805
|
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr11_-_41966854 | 2.55 |
ENSDART00000055709
|
her2
|
hairy-related 2 |
| chr6_-_40352215 | 2.55 |
ENSDART00000103992
|
ttll3
|
tubulin tyrosine ligase-like family, member 3 |
| chr19_+_2877079 | 2.53 |
ENSDART00000160847
|
hgh1
|
HGH1 homolog (S. cerevisiae) |
| chr7_-_6357952 | 2.50 |
ENSDART00000173197
|
zgc:165555
|
zgc:165555 |
| chr13_-_24260609 | 2.44 |
ENSDART00000138747
|
urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr4_-_685412 | 2.41 |
ENSDART00000168167
|
rtcb
|
RNA 2',3'-cyclic phosphate and 5'-OH ligase |
| chr6_-_39344259 | 2.39 |
ENSDART00000104074
|
zgc:158846
|
zgc:158846 |
| chr7_-_66864756 | 2.39 |
ENSDART00000184462
ENSDART00000189424 |
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr17_+_4368859 | 2.38 |
ENSDART00000055385
|
crls1
|
cardiolipin synthase 1 |
| chr15_+_32727848 | 2.36 |
ENSDART00000161361
|
postnb
|
periostin, osteoblast specific factor b |
| chr1_-_20593778 | 2.34 |
ENSDART00000124770
|
ugt8
|
UDP glycosyltransferase 8 |
| chr23_-_44470253 | 2.30 |
ENSDART00000176333
|
si:ch1073-228j22.2
|
si:ch1073-228j22.2 |
| chr11_+_21096339 | 2.27 |
ENSDART00000124574
|
il19l
|
interleukin 19 like |
| chr1_+_56843411 | 2.21 |
ENSDART00000152312
|
si:ch211-152f2.1
|
si:ch211-152f2.1 |
| chr17_-_14836320 | 2.20 |
ENSDART00000157051
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr7_-_17690983 | 2.16 |
ENSDART00000009367
|
map4k2
|
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr7_-_24047316 | 2.16 |
ENSDART00000184799
|
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr8_-_25247284 | 2.14 |
ENSDART00000132697
|
gnat2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 |
| chr24_-_25166720 | 2.13 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr22_-_10470663 | 2.13 |
ENSDART00000143352
|
omd
|
osteomodulin |
| chr22_-_4798738 | 2.12 |
ENSDART00000114465
|
zgc:195170
|
zgc:195170 |
| chr24_-_11905911 | 2.11 |
ENSDART00000033621
|
tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr9_-_39624173 | 2.10 |
ENSDART00000180106
ENSDART00000126766 |
erbb4b
|
erb-b2 receptor tyrosine kinase 4b |
| chr8_+_14058646 | 2.05 |
ENSDART00000080852
|
ugt5e1
|
UDP glucuronosyltransferase 5 family, polypeptide E1 |
| chr12_+_20693743 | 2.03 |
ENSDART00000153023
ENSDART00000153370 |
st6galnac1.2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1, tandem duplicate 2 |
| chr21_-_12749501 | 2.02 |
ENSDART00000179724
|
LO018011.1
|
|
| chr10_-_7892192 | 2.01 |
ENSDART00000145480
|
smtna
|
smoothelin a |
| chr10_+_24445698 | 1.98 |
ENSDART00000146370
|
slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr3_-_41715690 | 1.97 |
ENSDART00000184703
|
BX324110.1
|
|
| chr6_-_8360918 | 1.97 |
ENSDART00000004716
|
acp5a
|
acid phosphatase 5a, tartrate resistant |
| chr17_+_38030327 | 1.95 |
ENSDART00000085481
|
slc25a21
|
solute carrier family 25 (mitochondrial oxoadipate carrier), member 21 |
| chr10_+_26935155 | 1.94 |
ENSDART00000129993
|
CR450793.1
|
|
| chr10_+_35491216 | 1.93 |
ENSDART00000109705
|
phldb2a
|
pleckstrin homology-like domain, family B, member 2a |
| chr6_-_31224563 | 1.89 |
ENSDART00000104616
|
lepr
|
leptin receptor |
| chr21_+_21357309 | 1.88 |
ENSDART00000079634
ENSDART00000146102 |
kcnk12l
|
potassium channel, subfamily K, member 12 like |
| chr18_+_33132266 | 1.87 |
ENSDART00000151623
|
si:ch211-229c8.14
|
si:ch211-229c8.14 |
| chr16_-_1709328 | 1.85 |
ENSDART00000168865
|
ascc3
|
activating signal cointegrator 1 complex subunit 3 |
| chr5_-_64900552 | 1.85 |
ENSDART00000073963
|
si:ch211-236k19.2
|
si:ch211-236k19.2 |
| chr5_+_23630384 | 1.83 |
ENSDART00000013745
|
cx39.9
|
connexin 39.9 |
| chr22_-_22231720 | 1.80 |
ENSDART00000160165
|
ap3d1
|
adaptor-related protein complex 3, delta 1 subunit |
| chr22_-_37738203 | 1.80 |
ENSDART00000143190
|
acap2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr17_+_8799451 | 1.79 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr9_+_3519191 | 1.77 |
ENSDART00000008606
|
mettl8
|
methyltransferase like 8 |
| chr14_+_5835134 | 1.77 |
ENSDART00000054867
|
aup1
|
ancient ubiquitous protein 1 |
| chr5_+_50879545 | 1.75 |
ENSDART00000128402
|
nol6
|
nucleolar protein 6 (RNA-associated) |
| chr25_-_24216980 | 1.73 |
ENSDART00000135349
|
si:dkeyp-19e1.4
|
si:dkeyp-19e1.4 |
| chr15_-_18197008 | 1.69 |
ENSDART00000147632
|
si:ch211-247l8.8
|
si:ch211-247l8.8 |
| chr22_-_10891213 | 1.68 |
ENSDART00000145229
|
arhgef18b
|
rho/rac guanine nucleotide exchange factor (GEF) 18b |
| chr14_+_80685 | 1.67 |
ENSDART00000188443
|
stag3
|
stromal antigen 3 |
| chr4_-_18635005 | 1.63 |
ENSDART00000125361
|
pparaa
|
peroxisome proliferator-activated receptor alpha a |
| chr5_+_58455488 | 1.63 |
ENSDART00000038602
ENSDART00000127958 |
slc37a2
|
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr13_-_280827 | 1.61 |
ENSDART00000144819
|
slc30a6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr18_-_26797723 | 1.61 |
ENSDART00000008013
|
sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr1_-_34450784 | 1.60 |
ENSDART00000140515
|
lmo7b
|
LIM domain 7b |
| chr19_+_31541607 | 1.60 |
ENSDART00000181604
|
gmnn
|
geminin, DNA replication inhibitor |
| chr2_+_30481125 | 1.59 |
ENSDART00000125933
|
fam173b
|
family with sequence similarity 173, member B |
| chr13_+_11876145 | 1.59 |
ENSDART00000128249
|
trim8a
|
tripartite motif containing 8a |
| chr1_-_34450622 | 1.58 |
ENSDART00000083736
|
lmo7b
|
LIM domain 7b |
| chr25_-_19655820 | 1.57 |
ENSDART00000149585
ENSDART00000104353 |
atp2b1b
|
ATPase plasma membrane Ca2+ transporting 1b |
| chr24_-_35282568 | 1.57 |
ENSDART00000167406
ENSDART00000088609 |
sntg1
|
syntrophin, gamma 1 |
| chr7_+_30051880 | 1.57 |
ENSDART00000075609
|
pstpip1b
|
proline-serine-threonine phosphatase interacting protein 1b |
| chr24_+_4434117 | 1.56 |
ENSDART00000184349
|
GJD4
|
gap junction protein delta 4 |
| chr14_-_26465729 | 1.56 |
ENSDART00000143454
|
syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr16_-_42750295 | 1.55 |
ENSDART00000176570
|
slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr20_-_42439896 | 1.51 |
ENSDART00000061049
|
vgll2a
|
vestigial-like family member 2a |
| chr18_+_34861568 | 1.50 |
ENSDART00000192825
|
LO018333.1
|
|
| chr20_-_18789543 | 1.50 |
ENSDART00000182240
|
ccm2
|
cerebral cavernous malformation 2 |
| chr2_-_31833347 | 1.49 |
ENSDART00000109460
|
nol7
|
nucleolar protein 7 |
| chr7_+_1473929 | 1.49 |
ENSDART00000050687
|
lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
| chr3_-_12227359 | 1.48 |
ENSDART00000167356
|
tfap4
|
transcription factor AP-4 (activating enhancer binding protein 4) |
| chr4_-_17680493 | 1.48 |
ENSDART00000180131
|
BX890572.2
|
|
| chr8_+_30456161 | 1.43 |
ENSDART00000085894
|
pgm5
|
phosphoglucomutase 5 |
| chr16_-_12291045 | 1.43 |
ENSDART00000185447
|
CR388102.1
|
|
| chr12_-_9294819 | 1.40 |
ENSDART00000003805
|
pth1rb
|
parathyroid hormone 1 receptor b |
| chr7_+_27277236 | 1.40 |
ENSDART00000185161
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr18_-_10995410 | 1.40 |
ENSDART00000136751
|
tspan33b
|
tetraspanin 33b |
| chr7_+_4971938 | 1.39 |
ENSDART00000172889
ENSDART00000145389 |
si:dkey-81n2.2
|
si:dkey-81n2.2 |
| chr1_-_40911332 | 1.38 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr5_-_24238733 | 1.38 |
ENSDART00000138170
|
plscr3a
|
phospholipid scramblase 3a |
| chr12_+_2660733 | 1.38 |
ENSDART00000152235
|
irbpl
|
interphotoreceptor retinoid-binding protein like |
| chr12_+_30360184 | 1.36 |
ENSDART00000190718
|
si:ch211-225b10.3
|
si:ch211-225b10.3 |
| chr11_-_308838 | 1.35 |
ENSDART00000112538
|
poc1a
|
POC1 centriolar protein A |
| chr6_-_39218609 | 1.35 |
ENSDART00000133305
|
os9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr4_-_43374443 | 1.33 |
ENSDART00000191022
|
BX927253.3
|
|
| chr3_+_58379450 | 1.33 |
ENSDART00000155759
|
sdr42e2
|
short chain dehydrogenase/reductase family 42E, member 2 |
| chr16_+_47428721 | 1.30 |
ENSDART00000180597
ENSDART00000032309 |
slc25a32b
|
solute carrier family 25 (mitochondrial folate carrier), member 32b |
| chr2_+_29257942 | 1.29 |
ENSDART00000184362
ENSDART00000025562 |
cdh18a
|
cadherin 18, type 2a |
| chr15_+_37954666 | 1.28 |
ENSDART00000126534
ENSDART00000155548 |
si:dkey-238d18.7
|
si:dkey-238d18.7 |
| chr13_+_2625150 | 1.25 |
ENSDART00000164177
|
plpp4
|
phospholipid phosphatase 4 |
| chr5_-_57480660 | 1.25 |
ENSDART00000147875
ENSDART00000142776 |
si:ch211-202f5.2
|
si:ch211-202f5.2 |
| chr5_+_60886103 | 1.24 |
ENSDART00000009298
|
tmem248
|
transmembrane protein 248 |
| chr5_-_23729590 | 1.24 |
ENSDART00000014831
|
si:dkey-110k5.10
|
si:dkey-110k5.10 |
| chr14_+_34952883 | 1.22 |
ENSDART00000182812
|
il12ba
|
interleukin 12Ba |
| chr1_+_20593653 | 1.22 |
ENSDART00000132440
|
si:ch211-142c4.1
|
si:ch211-142c4.1 |
| chr13_+_21601149 | 1.20 |
ENSDART00000179369
|
sh2d4ba
|
SH2 domain containing 4Ba |
| chr6_+_37752781 | 1.20 |
ENSDART00000154364
|
herc2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr18_+_19820008 | 1.20 |
ENSDART00000079691
ENSDART00000135049 |
iqch
|
IQ motif containing H |
| chr22_+_14102397 | 1.20 |
ENSDART00000146397
ENSDART00000137530 |
he1.3
|
hatching enzyme 1, tandem duplicate 3 |
| chr21_+_39185461 | 1.20 |
ENSDART00000178419
|
cryba1b
|
crystallin, beta A1b |
| chr8_+_22405477 | 1.20 |
ENSDART00000148267
|
si:dkey-23c22.7
|
si:dkey-23c22.7 |
| chr20_+_18163355 | 1.20 |
ENSDART00000011287
|
aqp4
|
aquaporin 4 |
| chr15_-_945804 | 1.20 |
ENSDART00000063257
|
alox5b.1
|
arachidonate 5-lipoxygenase b, tandem duplicate 1 |
| chr21_-_11632403 | 1.19 |
ENSDART00000171708
ENSDART00000138619 ENSDART00000136308 ENSDART00000144770 |
cast
|
calpastatin |
| chr10_+_1590604 | 1.18 |
ENSDART00000166646
|
ora5
|
olfactory receptor class A related 5 |
| chr6_+_25261297 | 1.17 |
ENSDART00000162824
ENSDART00000163490 ENSDART00000157790 ENSDART00000160978 ENSDART00000161545 ENSDART00000159978 |
kyat3
|
kynurenine aminotransferase 3 |
| chr14_+_7939398 | 1.16 |
ENSDART00000189773
|
cxxc5b
|
CXXC finger protein 5b |
| chr2_+_17556177 | 1.16 |
ENSDART00000074113
|
pimr197
|
Pim proto-oncogene, serine/threonine kinase, related 197 |
| chr5_-_24008997 | 1.16 |
ENSDART00000066645
|
eif1axa
|
eukaryotic translation initiation factor 1A, X-linked, a |
| chr2_+_44977889 | 1.16 |
ENSDART00000144024
|
alg3
|
asparagine-linked glycosylation 3 (alpha-1,3-mannosyltransferase) |
| chr5_-_65121747 | 1.15 |
ENSDART00000165556
|
tor2a
|
torsin family 2, member A |
| chr25_+_36045072 | 1.15 |
ENSDART00000126326
|
rpgrip1l
|
RPGRIP1-like |
| chr21_-_39253647 | 1.14 |
ENSDART00000133034
|
tusc5b
|
tumor suppressor candidate 5b |
| chr11_-_11266882 | 1.14 |
ENSDART00000020256
|
lgsn
|
lengsin, lens protein with glutamine synthetase domain |
| chr4_-_21466480 | 1.12 |
ENSDART00000139962
|
pawr
|
PRKC, apoptosis, WT1, regulator |
| chr15_-_38009344 | 1.12 |
ENSDART00000157094
|
si:dkey-238d18.6
|
si:dkey-238d18.6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of tcf21
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 19.2 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 4.4 | 13.3 | GO:0032369 | negative regulation of lipid transport(GO:0032369) negative regulation of lipid catabolic process(GO:0050995) |
| 3.5 | 10.4 | GO:0015695 | organic cation transport(GO:0015695) |
| 1.9 | 11.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 1.5 | 7.7 | GO:0043011 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 1.5 | 5.9 | GO:0015860 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 1.4 | 124.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 1.2 | 6.2 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 1.0 | 4.2 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 1.0 | 3.1 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 1.0 | 6.0 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 1.0 | 7.7 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.9 | 5.7 | GO:1902946 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.9 | 5.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.9 | 12.6 | GO:1903963 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.7 | 5.7 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.7 | 2.7 | GO:0003400 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.6 | 7.5 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.6 | 1.8 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.5 | 1.6 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.4 | 1.3 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.4 | 4.5 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.4 | 2.7 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.4 | 2.6 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.4 | 2.6 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.4 | 4.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.4 | 2.6 | GO:0098815 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.3 | 1.0 | GO:0010658 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.3 | 2.4 | GO:0032049 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.3 | 4.6 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.3 | 2.8 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.2 | 8.7 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.2 | 1.8 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.2 | 1.8 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.2 | 1.8 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.2 | 5.0 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.2 | 2.2 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.2 | 3.3 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.2 | 11.8 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.2 | 2.1 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.2 | 2.7 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.2 | 0.9 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.2 | 1.6 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.2 | 2.0 | GO:0045453 | bone resorption(GO:0045453) |
| 0.2 | 5.4 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.2 | 1.6 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.2 | 1.7 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.2 | 0.5 | GO:0060148 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.2 | 0.5 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.1 | 1.4 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 8.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 1.2 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 3.7 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 0.9 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 1.4 | GO:0030656 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.1 | 1.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.8 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.2 | GO:2000009 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 1.2 | GO:0043651 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.6 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 0.7 | GO:0090179 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 1.8 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 4.9 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.1 | 0.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 1.9 | GO:0009749 | response to glucose(GO:0009749) |
| 0.1 | 1.1 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.1 | 1.6 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 2.1 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.1 | 1.9 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 1.1 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.1 | 0.5 | GO:0032475 | otolith formation(GO:0032475) |
| 0.1 | 4.6 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.1 | 5.2 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.1 | 1.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.5 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 0.8 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 1.0 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 2.2 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 0.8 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 0.7 | GO:0032196 | transposition(GO:0032196) |
| 0.1 | 2.4 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 1.1 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 0.5 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 0.9 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 8.8 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.1 | 4.4 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.2 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.0 | 1.4 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 7.4 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 2.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.5 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.2 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.0 | 0.5 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 1.7 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 1.8 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 1.5 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 1.1 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.0 | 1.1 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 0.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 2.5 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 6.0 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 1.7 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 2.6 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 5.0 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 3.2 | GO:0001947 | heart looping(GO:0001947) |
| 0.0 | 0.5 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.6 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.1 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 1.4 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 3.6 | GO:0006631 | fatty acid metabolic process(GO:0006631) |
| 0.0 | 1.3 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 0.4 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 1.4 | GO:0051604 | protein maturation(GO:0051604) |
| 0.0 | 1.5 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.9 | GO:0051170 | nuclear import(GO:0051170) |
| 0.0 | 0.3 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) dendritic spine organization(GO:0097061) |
| 0.0 | 0.0 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 1.1 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 1.2 | GO:0006413 | translational initiation(GO:0006413) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 13.3 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.7 | 5.7 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.6 | 1.8 | GO:0034456 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.6 | 3.3 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.5 | 6.7 | GO:0002102 | podosome(GO:0002102) |
| 0.4 | 13.0 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.4 | 1.6 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.3 | 2.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.3 | 1.2 | GO:0070743 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.3 | 4.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.3 | 5.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.3 | 1.8 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.3 | 1.8 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 2.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 1.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.2 | 6.2 | GO:0043186 | P granule(GO:0043186) |
| 0.2 | 7.5 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 2.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 3.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 5.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 2.8 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 134.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 11.0 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 2.1 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 2.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 2.1 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 6.0 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.1 | 1.6 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 9.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.7 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.1 | 0.6 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 2.9 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 2.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 2.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 5.7 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.3 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 1.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 13.1 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 2.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.8 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 3.3 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 7.9 | GO:1990904 | ribonucleoprotein complex(GO:1990904) |
| 0.0 | 1.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 11.4 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 2.1 | 6.2 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 1.5 | 13.5 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 1.5 | 7.5 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 1.5 | 5.9 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 1.0 | 13.3 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 1.0 | 118.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.9 | 2.6 | GO:0042806 | fucose binding(GO:0042806) |
| 0.9 | 10.4 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.9 | 6.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.9 | 9.5 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.8 | 2.5 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.8 | 5.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.8 | 4.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.6 | 5.7 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.5 | 1.6 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.5 | 11.8 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.4 | 1.8 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.4 | 1.3 | GO:0015230 | folic acid transporter activity(GO:0008517) FAD transmembrane transporter activity(GO:0015230) |
| 0.4 | 5.5 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.4 | 3.1 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.4 | 9.5 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.4 | 5.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.4 | 2.6 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.4 | 1.8 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.3 | 1.5 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.3 | 1.2 | GO:0019972 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.3 | 1.8 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.3 | 1.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.3 | 7.7 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.3 | 3.8 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.3 | 5.9 | GO:0010181 | FMN binding(GO:0010181) |
| 0.3 | 4.5 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.2 | 2.0 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.2 | 5.6 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.2 | 6.0 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 2.8 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.2 | 8.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 7.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 5.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 1.2 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.2 | 1.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.2 | 0.9 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 1.3 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 1.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 1.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 1.4 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 1.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.8 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 6.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 1.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.2 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.1 | 1.8 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 6.3 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 5.0 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 2.7 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 0.5 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 2.4 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.1 | 2.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 16.6 | GO:0008757 | S-adenosylmethionine-dependent methyltransferase activity(GO:0008757) |
| 0.1 | 1.1 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.8 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 4.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.9 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.1 | 0.9 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.9 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 1.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 1.4 | GO:0019840 | retinoid binding(GO:0005501) isoprenoid binding(GO:0019840) |
| 0.1 | 1.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.9 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 4.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 3.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 2.0 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 2.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 1.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 7.6 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 2.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 3.5 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 1.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 2.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.8 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 2.8 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 2.5 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 1.0 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 4.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 6.0 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.5 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 12.2 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 1.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.3 | 44.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.3 | 13.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.2 | 7.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 5.1 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 1.8 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 2.1 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 1.9 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 1.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 7.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 5.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 1.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 1.1 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 1.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 5.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 2.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 3.2 | 12.6 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 1.5 | 7.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 1.1 | 11.8 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.6 | 3.9 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.6 | 3.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.4 | 4.9 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.4 | 5.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.4 | 1.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 4.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.3 | 3.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 2.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 1.6 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.2 | 2.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 31.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.2 | 1.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 5.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.6 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 7.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 2.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 0.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.8 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.1 | 0.5 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 1.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.1 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.4 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |