Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
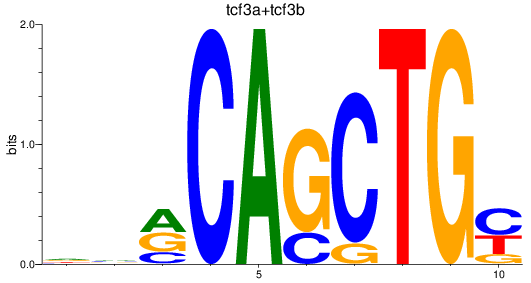
Results for tcf3a+tcf3b
Z-value: 1.15
Transcription factors associated with tcf3a+tcf3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tcf3a
|
ENSDARG00000005915 | transcription factor 3a |
|
tcf3b
|
ENSDARG00000099999 | transcription factor 3b |
|
tcf3b
|
ENSDARG00000112646 | transcription factor 3b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tcf3b | dr11_v1_chr22_-_20342260_20342260 | -0.33 | 1.2e-03 | Click! |
| tcf3a | dr11_v1_chr2_-_57378748_57378748 | -0.08 | 4.6e-01 | Click! |
Activity profile of tcf3a+tcf3b motif
Sorted Z-values of tcf3a+tcf3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_23647078 | 13.63 |
ENSDART00000059366
|
ckmb
|
creatine kinase, muscle b |
| chr22_+_11756040 | 13.57 |
ENSDART00000105808
|
krt97
|
keratin 97 |
| chr5_-_36837846 | 12.57 |
ENSDART00000032481
|
ckma
|
creatine kinase, muscle a |
| chr6_-_42003780 | 11.67 |
ENSDART00000032527
|
cav3
|
caveolin 3 |
| chr18_-_6634424 | 10.74 |
ENSDART00000062423
ENSDART00000179955 |
tnni1c
|
troponin I, skeletal, slow c |
| chr21_+_27382893 | 10.12 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr18_+_5454341 | 9.57 |
ENSDART00000192649
|
dtwd1
|
DTW domain containing 1 |
| chr6_-_15653494 | 8.24 |
ENSDART00000038133
|
trim63a
|
tripartite motif containing 63a |
| chr17_+_27434626 | 8.04 |
ENSDART00000052446
|
vgll2b
|
vestigial-like family member 2b |
| chr12_-_26064105 | 8.04 |
ENSDART00000168825
|
ldb3b
|
LIM domain binding 3b |
| chr8_-_1051438 | 8.03 |
ENSDART00000067093
ENSDART00000170737 |
smyd1b
|
SET and MYND domain containing 1b |
| chr13_+_22264914 | 7.78 |
ENSDART00000060576
|
myoz1a
|
myozenin 1a |
| chr2_-_21352101 | 7.66 |
ENSDART00000057021
|
hhatla
|
hedgehog acyltransferase like, a |
| chr14_+_31651533 | 7.66 |
ENSDART00000172835
|
fhl1a
|
four and a half LIM domains 1a |
| chr9_-_100579 | 7.62 |
ENSDART00000006099
|
lims2
|
LIM and senescent cell antigen-like domains 2 |
| chr23_-_5683147 | 7.48 |
ENSDART00000102766
ENSDART00000067351 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr24_+_39108243 | 7.26 |
ENSDART00000156353
|
mss51
|
MSS51 mitochondrial translational activator |
| chr22_+_11775269 | 7.22 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr13_-_438705 | 7.18 |
ENSDART00000082142
|
CU570800.1
|
|
| chr10_+_33171501 | 7.15 |
ENSDART00000159666
|
myl10
|
myosin, light chain 10, regulatory |
| chr7_+_29951997 | 7.07 |
ENSDART00000173453
|
tpma
|
alpha-tropomyosin |
| chr2_+_55982940 | 6.99 |
ENSDART00000097753
ENSDART00000097751 |
nmrk2
|
nicotinamide riboside kinase 2 |
| chr15_-_29387446 | 6.90 |
ENSDART00000145976
ENSDART00000035096 |
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr21_+_5129513 | 6.85 |
ENSDART00000102572
|
thbs4b
|
thrombospondin 4b |
| chr15_+_33989181 | 6.72 |
ENSDART00000169487
|
vwde
|
von Willebrand factor D and EGF domains |
| chr24_-_33703504 | 6.66 |
ENSDART00000079292
|
cavin4b
|
caveolae associated protein 4b |
| chr21_-_45882643 | 6.50 |
ENSDART00000168703
|
galnt10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) |
| chr7_+_31891110 | 6.48 |
ENSDART00000173883
|
mybpc3
|
myosin binding protein C, cardiac |
| chr22_-_14128716 | 6.35 |
ENSDART00000140323
|
si:ch211-246m6.4
|
si:ch211-246m6.4 |
| chr14_-_25956804 | 6.24 |
ENSDART00000135627
ENSDART00000146022 ENSDART00000039660 |
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr2_-_42128714 | 6.22 |
ENSDART00000047055
|
trim55a
|
tripartite motif containing 55a |
| chr2_-_44255537 | 6.11 |
ENSDART00000011188
ENSDART00000093298 |
atp1a2a
|
ATPase Na+/K+ transporting subunit alpha 2 |
| chr22_+_16308806 | 6.11 |
ENSDART00000162685
|
lrrc39
|
leucine rich repeat containing 39 |
| chr17_+_1323699 | 5.96 |
ENSDART00000172540
|
adssl1
|
adenylosuccinate synthase like 1 |
| chr11_-_30636163 | 5.94 |
ENSDART00000140516
|
zgc:153665
|
zgc:153665 |
| chr11_-_45171139 | 5.86 |
ENSDART00000167036
ENSDART00000161712 ENSDART00000158156 |
syngr2b
|
synaptogyrin 2b |
| chr22_+_18469004 | 5.83 |
ENSDART00000061430
|
cilp2
|
cartilage intermediate layer protein 2 |
| chr22_+_16308450 | 5.70 |
ENSDART00000105678
|
lrrc39
|
leucine rich repeat containing 39 |
| chr7_+_31838320 | 5.58 |
ENSDART00000144679
ENSDART00000174217 ENSDART00000122506 |
mybpc3
|
myosin binding protein C, cardiac |
| chr14_+_31657412 | 5.58 |
ENSDART00000105767
|
fhl1a
|
four and a half LIM domains 1a |
| chr19_-_35035857 | 5.57 |
ENSDART00000103253
|
bmp8a
|
bone morphogenetic protein 8a |
| chr6_-_21492752 | 5.53 |
ENSDART00000006843
ENSDART00000171479 |
cacng1a
|
calcium channel, voltage-dependent, gamma subunit 1a |
| chr1_+_55002583 | 5.51 |
ENSDART00000037250
|
si:ch211-196h16.12
|
si:ch211-196h16.12 |
| chr9_-_98982 | 5.48 |
ENSDART00000147882
|
lims2
|
LIM and senescent cell antigen-like domains 2 |
| chr9_-_105135 | 5.26 |
ENSDART00000180126
|
FQ377903.3
|
|
| chr24_-_9300160 | 5.23 |
ENSDART00000152378
|
tgif1
|
TGFB-induced factor homeobox 1 |
| chr1_+_55137943 | 5.22 |
ENSDART00000138070
ENSDART00000150510 ENSDART00000133472 ENSDART00000136378 |
mb
|
myoglobin |
| chr9_-_22892838 | 5.21 |
ENSDART00000143888
|
neb
|
nebulin |
| chr1_-_50710468 | 5.04 |
ENSDART00000080389
|
fam13a
|
family with sequence similarity 13, member A |
| chr6_+_3680651 | 5.03 |
ENSDART00000013588
|
klhl41b
|
kelch-like family member 41b |
| chr23_-_10177442 | 4.94 |
ENSDART00000144280
ENSDART00000129044 |
krt5
|
keratin 5 |
| chr23_-_39666519 | 4.93 |
ENSDART00000110868
ENSDART00000190961 |
vwa1
|
von Willebrand factor A domain containing 1 |
| chr18_+_29145681 | 4.84 |
ENSDART00000089031
ENSDART00000193336 |
ppfibp2a
|
PTPRF interacting protein, binding protein 2a (liprin beta 2) |
| chr9_-_48281941 | 4.79 |
ENSDART00000099787
|
klhl41a
|
kelch-like family member 41a |
| chr22_-_651719 | 4.79 |
ENSDART00000148692
|
apobec2a
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2a |
| chr4_-_14915268 | 4.66 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
| chr8_+_25767610 | 4.65 |
ENSDART00000062406
|
cacna1sb
|
calcium channel, voltage-dependent, L type, alpha 1S subunit, b |
| chr13_+_22249636 | 4.64 |
ENSDART00000108472
ENSDART00000173123 |
synpo2la
|
synaptopodin 2-like a |
| chr23_-_45405968 | 4.62 |
ENSDART00000149462
|
zgc:101853
|
zgc:101853 |
| chr2_+_55982300 | 4.61 |
ENSDART00000183903
|
nmrk2
|
nicotinamide riboside kinase 2 |
| chr6_-_35401282 | 4.60 |
ENSDART00000127612
|
rgs5a
|
regulator of G protein signaling 5a |
| chr3_-_50865079 | 4.60 |
ENSDART00000164295
|
pmp22a
|
peripheral myelin protein 22a |
| chr18_-_26101800 | 4.58 |
ENSDART00000004692
|
idh2
|
isocitrate dehydrogenase 2 (NADP+), mitochondrial |
| chr6_-_21189295 | 4.57 |
ENSDART00000137136
|
obsl1a
|
obscurin-like 1a |
| chr7_+_69841017 | 4.55 |
ENSDART00000169107
|
FO818704.1
|
|
| chr19_-_25519612 | 4.55 |
ENSDART00000133150
|
C1GALT1 (1 of many)
|
si:dkey-202e17.1 |
| chr16_-_14074594 | 4.54 |
ENSDART00000090234
|
trim109
|
tripartite motif containing 109 |
| chr20_+_46040666 | 4.47 |
ENSDART00000060744
|
si:dkey-7c18.24
|
si:dkey-7c18.24 |
| chr11_-_18705303 | 4.40 |
ENSDART00000059732
|
id1
|
inhibitor of DNA binding 1 |
| chr23_+_1730663 | 4.36 |
ENSDART00000149545
|
tgm1
|
transglutaminase 1, K polypeptide |
| chr7_+_22767678 | 4.35 |
ENSDART00000137203
|
ponzr6
|
plac8 onzin related protein 6 |
| chr6_-_35439406 | 4.33 |
ENSDART00000073784
|
rgs5a
|
regulator of G protein signaling 5a |
| chr4_-_4834347 | 4.29 |
ENSDART00000141803
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr9_+_307863 | 4.25 |
ENSDART00000163474
|
stac3
|
SH3 and cysteine rich domain 3 |
| chr24_+_20559009 | 4.24 |
ENSDART00000131677
|
hhatlb
|
hedgehog acyltransferase like, b |
| chr23_-_31512496 | 4.21 |
ENSDART00000158755
ENSDART00000143425 |
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr13_-_46200240 | 4.17 |
ENSDART00000056984
|
ftr69
|
finTRIM family, member 69 |
| chr20_-_38758797 | 4.13 |
ENSDART00000061394
|
trim54
|
tripartite motif containing 54 |
| chr19_-_2861444 | 4.13 |
ENSDART00000169053
|
clec3bb
|
C-type lectin domain family 3, member Bb |
| chr22_-_57177 | 4.12 |
ENSDART00000163959
|
CABZ01085139.1
|
|
| chr7_+_35229805 | 4.12 |
ENSDART00000173911
|
tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr3_-_30685401 | 4.11 |
ENSDART00000151097
|
si:ch211-51c14.1
|
si:ch211-51c14.1 |
| chr20_+_23501535 | 4.10 |
ENSDART00000177922
ENSDART00000058532 |
palld
|
palladin, cytoskeletal associated protein |
| chr5_+_15203421 | 4.10 |
ENSDART00000040826
|
tbx1
|
T-box 1 |
| chr23_+_41889093 | 4.10 |
ENSDART00000138132
|
podn
|
podocan |
| chr19_-_25519310 | 4.09 |
ENSDART00000089882
|
C1GALT1 (1 of many)
|
si:dkey-202e17.1 |
| chr21_+_6556635 | 4.06 |
ENSDART00000139598
|
col5a1
|
procollagen, type V, alpha 1 |
| chr14_-_17659420 | 4.05 |
ENSDART00000165566
|
si:ch211-159i8.4
|
si:ch211-159i8.4 |
| chr12_-_36268723 | 4.04 |
ENSDART00000113740
|
kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr16_-_17713859 | 4.01 |
ENSDART00000149275
|
zgc:174935
|
zgc:174935 |
| chr13_-_37615029 | 4.01 |
ENSDART00000111199
|
si:dkey-188i13.6
|
si:dkey-188i13.6 |
| chr9_-_23990416 | 4.00 |
ENSDART00000113176
|
col6a3
|
collagen, type VI, alpha 3 |
| chr18_-_49020066 | 3.99 |
ENSDART00000174394
|
BX663503.3
|
|
| chr17_-_681142 | 3.96 |
ENSDART00000165583
|
soul3
|
heme-binding protein soul3 |
| chr23_+_19213472 | 3.96 |
ENSDART00000185985
|
apobec2b
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2b |
| chr20_-_21672970 | 3.93 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr19_-_5372572 | 3.90 |
ENSDART00000151326
|
krt17
|
keratin 17 |
| chr22_-_3914162 | 3.90 |
ENSDART00000187174
ENSDART00000190612 ENSDART00000187928 ENSDART00000057224 ENSDART00000184758 |
mhc1uma
|
major histocompatibility complex class I UMA |
| chr9_-_30145080 | 3.84 |
ENSDART00000133746
|
abi3bpa
|
ABI family, member 3 (NESH) binding protein a |
| chr16_-_31469065 | 3.82 |
ENSDART00000182397
|
si:ch211-251p5.5
|
si:ch211-251p5.5 |
| chr3_-_34095221 | 3.80 |
ENSDART00000164235
ENSDART00000151377 |
ighv1-4
ighv5-4
|
immunoglobulin heavy variable 1-4 immunoglobulin heavy variable 5-4 |
| chr12_+_5708400 | 3.76 |
ENSDART00000017191
|
dlx3b
|
distal-less homeobox 3b |
| chr5_+_64900223 | 3.74 |
ENSDART00000191677
|
ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr22_-_10541372 | 3.72 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr4_-_16412084 | 3.70 |
ENSDART00000188460
|
dcn
|
decorin |
| chr19_+_30990815 | 3.70 |
ENSDART00000134645
|
sync
|
syncoilin, intermediate filament protein |
| chr14_-_41678357 | 3.69 |
ENSDART00000185925
|
fgfrl1b
|
fibroblast growth factor receptor like 1b |
| chr6_+_18142623 | 3.68 |
ENSDART00000169431
ENSDART00000158841 |
si:dkey-237i9.8
|
si:dkey-237i9.8 |
| chr7_+_65586624 | 3.67 |
ENSDART00000184344
|
mical2a
|
microtubule associated monooxygenase, calponin and LIM domain containing 2a |
| chr14_-_9281232 | 3.65 |
ENSDART00000054693
|
asb12b
|
ankyrin repeat and SOCS box-containing 12b |
| chr19_-_32804535 | 3.63 |
ENSDART00000175613
ENSDART00000052098 |
nt5c1aa
|
5'-nucleotidase, cytosolic IAa |
| chr13_+_228045 | 3.60 |
ENSDART00000161091
|
zgc:64201
|
zgc:64201 |
| chr8_+_7033049 | 3.59 |
ENSDART00000064172
ENSDART00000134440 |
gpd1a
|
glycerol-3-phosphate dehydrogenase 1a |
| chr13_-_349952 | 3.59 |
ENSDART00000133731
ENSDART00000140326 ENSDART00000189389 ENSDART00000185865 ENSDART00000109634 ENSDART00000147058 ENSDART00000142695 |
si:ch1073-291c23.2
|
si:ch1073-291c23.2 |
| chr22_+_3914318 | 3.57 |
ENSDART00000188774
ENSDART00000082034 |
FO904903.1
|
Danio rerio major histocompatibility complex class I ULA (mhc1ula), mRNA. |
| chr3_-_22242868 | 3.55 |
ENSDART00000184511
ENSDART00000191558 ENSDART00000179846 |
myl4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr14_+_38786298 | 3.53 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr12_+_6214041 | 3.53 |
ENSDART00000179759
|
prkg1b
|
protein kinase, cGMP-dependent, type Ib |
| chr8_-_11229523 | 3.50 |
ENSDART00000002164
|
unc45b
|
unc-45 myosin chaperone B |
| chr14_-_17563773 | 3.43 |
ENSDART00000082667
|
fgfrl1a
|
fibroblast growth factor receptor like 1a |
| chr23_+_6077503 | 3.41 |
ENSDART00000081714
ENSDART00000139834 |
mybpha
|
myosin binding protein Ha |
| chr3_-_1190132 | 3.39 |
ENSDART00000149709
|
smdt1a
|
single-pass membrane protein with aspartate-rich tail 1a |
| chr12_-_46228023 | 3.37 |
ENSDART00000153455
|
si:ch211-226h7.6
|
si:ch211-226h7.6 |
| chr23_-_32162810 | 3.37 |
ENSDART00000155905
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr24_+_26276805 | 3.35 |
ENSDART00000089749
|
adipoqa
|
adiponectin, C1Q and collagen domain containing, a |
| chr10_+_42358426 | 3.33 |
ENSDART00000025691
|
dbnla
|
drebrin-like a |
| chr12_-_46145635 | 3.33 |
ENSDART00000074682
|
zgc:153932
|
zgc:153932 |
| chr8_-_43923788 | 3.33 |
ENSDART00000146152
|
adgrd1
|
adhesion G protein-coupled receptor D1 |
| chr23_+_19790962 | 3.33 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr16_-_14353567 | 3.30 |
ENSDART00000139859
|
itga10
|
integrin, alpha 10 |
| chr19_+_19772765 | 3.29 |
ENSDART00000182028
ENSDART00000161019 |
hoxa3a
|
homeobox A3a |
| chr23_+_18722715 | 3.29 |
ENSDART00000137438
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr14_-_36412473 | 3.28 |
ENSDART00000128244
ENSDART00000138376 |
asb5a
|
ankyrin repeat and SOCS box containing 5a |
| chr9_-_34396264 | 3.28 |
ENSDART00000045754
|
grtp1b
|
growth hormone regulated TBC protein 1b |
| chr3_+_26135502 | 3.28 |
ENSDART00000146979
|
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr5_-_41831646 | 3.27 |
ENSDART00000134326
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr20_-_35578435 | 3.27 |
ENSDART00000142444
|
adgrf6
|
adhesion G protein-coupled receptor F6 |
| chr4_-_25064510 | 3.26 |
ENSDART00000025153
|
gata3
|
GATA binding protein 3 |
| chr23_+_43177290 | 3.26 |
ENSDART00000193300
ENSDART00000186065 |
si:dkey-65j6.2
|
si:dkey-65j6.2 |
| chr5_-_14521500 | 3.21 |
ENSDART00000176565
|
si:ch211-244o22.2
|
si:ch211-244o22.2 |
| chr12_-_25380028 | 3.21 |
ENSDART00000142674
|
zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr21_+_28747069 | 3.21 |
ENSDART00000014058
|
zgc:100829
|
zgc:100829 |
| chr16_+_12236339 | 3.19 |
ENSDART00000132468
|
tpi1b
|
triosephosphate isomerase 1b |
| chr17_+_132555 | 3.19 |
ENSDART00000158159
|
zgc:77287
|
zgc:77287 |
| chr5_+_26212621 | 3.19 |
ENSDART00000134432
|
oclnb
|
occludin b |
| chr4_-_858434 | 3.17 |
ENSDART00000006961
|
sobpb
|
sine oculis binding protein homolog (Drosophila) b |
| chr1_-_10841348 | 3.15 |
ENSDART00000148305
|
dmd
|
dystrophin |
| chr19_+_24488403 | 3.14 |
ENSDART00000052421
|
txnipa
|
thioredoxin interacting protein a |
| chr3_-_32590164 | 3.13 |
ENSDART00000151151
|
tspan4b
|
tetraspanin 4b |
| chr19_+_5640504 | 3.13 |
ENSDART00000179987
|
ft2
|
alpha(1,3)fucosyltransferase gene 2 |
| chr18_+_36631923 | 3.13 |
ENSDART00000098980
|
znf296
|
zinc finger protein 296 |
| chr7_+_35229645 | 3.13 |
ENSDART00000144327
|
tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr24_+_5237753 | 3.12 |
ENSDART00000106488
ENSDART00000005901 |
plod2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr5_+_13373593 | 3.05 |
ENSDART00000051668
ENSDART00000183883 |
ccl19a.2
|
chemokine (C-C motif) ligand 19a, tandem duplicate 2 |
| chr3_+_13559199 | 3.03 |
ENSDART00000166547
|
si:ch73-106n3.1
|
si:ch73-106n3.1 |
| chr9_-_41784799 | 3.00 |
ENSDART00000144573
ENSDART00000112542 ENSDART00000190486 |
obsl1b
|
obscurin-like 1b |
| chr5_-_11271209 | 2.99 |
ENSDART00000122084
|
prodhb
|
proline dehydrogenase (oxidase) 1b |
| chr9_+_17309195 | 2.98 |
ENSDART00000048548
|
scel
|
sciellin |
| chr23_-_19686791 | 2.97 |
ENSDART00000161973
|
zgc:193598
|
zgc:193598 |
| chr21_+_28747236 | 2.95 |
ENSDART00000137874
|
zgc:100829
|
zgc:100829 |
| chr2_+_7106837 | 2.95 |
ENSDART00000138691
|
vcam1a
|
vascular cell adhesion molecule 1a |
| chr4_-_78026285 | 2.92 |
ENSDART00000168273
|
CCT2
|
chaperonin containing TCP1 subunit 2 |
| chr7_-_73843720 | 2.91 |
ENSDART00000111622
|
caap1
|
caspase activity and apoptosis inhibitor 1 |
| chr13_-_17723417 | 2.90 |
ENSDART00000183834
|
vdac2
|
voltage-dependent anion channel 2 |
| chr13_+_32144370 | 2.89 |
ENSDART00000020270
|
osr1
|
odd-skipped related transciption factor 1 |
| chr20_-_31067306 | 2.89 |
ENSDART00000014163
|
fndc1
|
fibronectin type III domain containing 1 |
| chr17_+_33433576 | 2.88 |
ENSDART00000077581
|
snap23.2
|
synaptosomal-associated protein 23.2 |
| chr19_-_10771558 | 2.88 |
ENSDART00000085165
|
slc9a3.2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3, tandem duplicate 2 |
| chr18_+_54354 | 2.87 |
ENSDART00000097163
|
zgc:158482
|
zgc:158482 |
| chr11_+_1845787 | 2.87 |
ENSDART00000173062
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr13_-_31622195 | 2.86 |
ENSDART00000057432
|
six1a
|
SIX homeobox 1a |
| chr23_+_18722915 | 2.86 |
ENSDART00000025057
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr6_+_2097690 | 2.86 |
ENSDART00000193770
|
tgm2b
|
transglutaminase 2b |
| chr21_-_20832482 | 2.86 |
ENSDART00000191928
|
c6
|
complement component 6 |
| chr17_-_25382367 | 2.86 |
ENSDART00000162306
ENSDART00000165282 |
lck
|
LCK proto-oncogene, Src family tyrosine kinase |
| chr17_+_38573471 | 2.85 |
ENSDART00000040627
|
sptb
|
spectrin, beta, erythrocytic |
| chr2_+_30379650 | 2.84 |
ENSDART00000129542
|
crispld1b
|
cysteine-rich secretory protein LCCL domain containing 1b |
| chr23_-_5759242 | 2.82 |
ENSDART00000055087
|
phlda3
|
pleckstrin homology-like domain, family A, member 3 |
| chr1_-_50247 | 2.82 |
ENSDART00000168428
|
ildr1a
|
immunoglobulin-like domain containing receptor 1a |
| chr20_+_46385907 | 2.82 |
ENSDART00000060710
|
adgrg11
|
adhesion G protein-coupled receptor G11 |
| chr14_+_146857 | 2.79 |
ENSDART00000122521
|
CABZ01088229.1
|
|
| chr1_-_38816685 | 2.78 |
ENSDART00000075230
|
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr20_-_49681850 | 2.77 |
ENSDART00000025926
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr2_-_23172708 | 2.76 |
ENSDART00000041365
|
prrx1a
|
paired related homeobox 1a |
| chr7_+_13756374 | 2.75 |
ENSDART00000180808
|
rasl12
|
RAS-like, family 12 |
| chr12_+_18524953 | 2.75 |
ENSDART00000090332
|
neurl2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr8_-_7093507 | 2.74 |
ENSDART00000045669
|
si:dkey-222n6.2
|
si:dkey-222n6.2 |
| chr18_+_2228737 | 2.73 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr7_-_41858513 | 2.72 |
ENSDART00000109918
|
mylk3
|
myosin light chain kinase 3 |
| chr21_-_38153824 | 2.71 |
ENSDART00000151226
|
klf5l
|
Kruppel-like factor 5 like |
| chr19_+_2685779 | 2.70 |
ENSDART00000160533
ENSDART00000097531 |
tomm7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr1_+_31054915 | 2.70 |
ENSDART00000148968
|
itga6b
|
integrin, alpha 6b |
| chr6_-_39051319 | 2.69 |
ENSDART00000155093
|
tns2b
|
tensin 2b |
| chr5_+_6670945 | 2.69 |
ENSDART00000185686
|
pxna
|
paxillin a |
| chr23_+_384850 | 2.68 |
ENSDART00000114000
|
zgc:101663
|
zgc:101663 |
| chr3_+_16722014 | 2.67 |
ENSDART00000008711
|
gys1
|
glycogen synthase 1 (muscle) |
| chr11_+_37096520 | 2.67 |
ENSDART00000181503
|
card19
|
caspase recruitment domain family, member 19 |
| chr11_+_21050326 | 2.66 |
ENSDART00000065984
|
zgc:113307
|
zgc:113307 |
| chr9_-_49531762 | 2.66 |
ENSDART00000121875
|
xirp2b
|
xin actin binding repeat containing 2b |
| chr3_-_48980319 | 2.66 |
ENSDART00000165319
|
ftr42
|
finTRIM family, member 42 |
Network of associatons between targets according to the STRING database.
First level regulatory network of tcf3a+tcf3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 11.9 | GO:1903060 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 2.9 | 8.7 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 2.5 | 32.8 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 2.0 | 6.1 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 2.0 | 26.2 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 1.8 | 5.3 | GO:0048389 | intermediate mesoderm development(GO:0048389) |
| 1.6 | 13.1 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 1.6 | 14.0 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 1.5 | 4.6 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 1.4 | 4.1 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 1.4 | 10.9 | GO:0033292 | T-tubule organization(GO:0033292) |
| 1.3 | 6.5 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 1.3 | 3.8 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 1.2 | 7.2 | GO:0035989 | tendon development(GO:0035989) |
| 1.1 | 5.5 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 1.0 | 2.1 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.9 | 3.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.9 | 5.4 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.9 | 3.6 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.9 | 5.2 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.8 | 2.5 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.8 | 3.3 | GO:0031448 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.8 | 8.0 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.8 | 3.2 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.8 | 3.1 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.8 | 3.1 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.8 | 2.3 | GO:0050787 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.7 | 2.2 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.7 | 2.9 | GO:0014857 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.7 | 2.1 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.7 | 3.3 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.7 | 4.0 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.7 | 5.3 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.6 | 4.5 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.6 | 9.8 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.6 | 1.8 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.6 | 3.0 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.6 | 2.3 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.6 | 10.1 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.6 | 2.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.5 | 4.4 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.5 | 11.8 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.5 | 3.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.5 | 1.5 | GO:0001977 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 0.5 | 1.4 | GO:0034138 | toll-like receptor 2 signaling pathway(GO:0034134) toll-like receptor 3 signaling pathway(GO:0034138) |
| 0.5 | 1.4 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) chiasma assembly(GO:0051026) |
| 0.5 | 4.2 | GO:1901099 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.5 | 1.4 | GO:0032060 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.5 | 3.3 | GO:0045582 | positive regulation of T cell differentiation(GO:0045582) |
| 0.5 | 2.7 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.4 | 1.3 | GO:0061182 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.4 | 2.5 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.4 | 1.7 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.4 | 2.0 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.4 | 3.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.4 | 26.2 | GO:0006958 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.4 | 12.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.4 | 1.8 | GO:0003010 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.4 | 1.1 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.4 | 2.9 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.4 | 2.1 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.3 | 2.8 | GO:0086002 | cardiac muscle cell action potential involved in contraction(GO:0086002) |
| 0.3 | 2.4 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.3 | 6.1 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.3 | 6.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.3 | 7.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.3 | 2.3 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.3 | 0.9 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.3 | 1.3 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.3 | 2.7 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.3 | 2.1 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.3 | 3.3 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.3 | 1.8 | GO:0050655 | dermatan sulfate proteoglycan metabolic process(GO:0050655) |
| 0.3 | 10.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.3 | 0.6 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.3 | 1.4 | GO:0090200 | regulation of mitochondrial membrane potential(GO:0051881) regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.3 | 4.6 | GO:0014823 | response to activity(GO:0014823) |
| 0.3 | 0.3 | GO:0001659 | temperature homeostasis(GO:0001659) |
| 0.3 | 1.1 | GO:0044060 | regulation of endocrine process(GO:0044060) endocrine hormone secretion(GO:0060986) |
| 0.3 | 1.9 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.3 | 2.5 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.3 | 3.2 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.3 | 1.5 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.3 | 0.8 | GO:0001113 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.2 | 1.5 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.2 | 2.7 | GO:0000022 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.2 | 2.5 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.2 | 1.2 | GO:1903573 | negative regulation of response to endoplasmic reticulum stress(GO:1903573) |
| 0.2 | 0.7 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.2 | 1.7 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 1.7 | GO:0035912 | aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.2 | 1.0 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.2 | 6.2 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.2 | 1.0 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 0.2 | 0.7 | GO:0090008 | convergent extension involved in nephron morphogenesis(GO:0072045) hypoblast development(GO:0090008) |
| 0.2 | 1.4 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.2 | 2.8 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 0.9 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.2 | 5.7 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 2.7 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.2 | 2.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 1.6 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.2 | 4.3 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.2 | 0.9 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.2 | 1.5 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.2 | 1.8 | GO:1901998 | toxin transport(GO:1901998) |
| 0.2 | 9.8 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.2 | 0.6 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.2 | 0.9 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 6.7 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.2 | 1.5 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.2 | 2.2 | GO:0033198 | response to ATP(GO:0033198) |
| 0.2 | 4.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 2.7 | GO:0061055 | myotome development(GO:0061055) |
| 0.2 | 3.7 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.2 | 2.7 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.2 | 1.6 | GO:0021794 | thalamus development(GO:0021794) |
| 0.2 | 1.0 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.2 | 1.2 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.2 | 1.6 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.2 | 0.9 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.2 | 1.0 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.2 | 4.3 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.2 | 10.5 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.2 | 1.2 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.2 | 2.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.2 | 0.8 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.2 | 2.6 | GO:0036065 | fucosylation(GO:0036065) |
| 0.2 | 0.5 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.2 | 1.9 | GO:2000406 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.2 | 0.6 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.2 | 4.4 | GO:0048794 | swim bladder development(GO:0048794) |
| 0.2 | 1.7 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.2 | 3.9 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 1.5 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 1.6 | GO:0001840 | neural plate development(GO:0001840) |
| 0.1 | 0.4 | GO:0051037 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.1 | 4.7 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 2.9 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 3.7 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 2.8 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.1 | 2.1 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 1.4 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.7 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 0.3 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 1.5 | GO:0070307 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 4.4 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 0.8 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.1 | 0.7 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.1 | 2.1 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 1.0 | GO:1902882 | regulation of cellular response to oxidative stress(GO:1900407) regulation of response to oxidative stress(GO:1902882) |
| 0.1 | 10.0 | GO:1990266 | neutrophil migration(GO:1990266) |
| 0.1 | 10.3 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 0.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.8 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.1 | 0.9 | GO:1905207 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.1 | 6.5 | GO:0072676 | lymphocyte migration(GO:0072676) |
| 0.1 | 10.2 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.1 | 2.5 | GO:0070672 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.1 | 1.9 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 0.7 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 2.2 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 1.0 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 1.3 | GO:0021684 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.4 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 3.1 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 0.5 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.1 | 1.8 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.3 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 5.2 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.1 | 0.3 | GO:0003343 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) |
| 0.1 | 2.0 | GO:0010634 | positive regulation of epithelial cell migration(GO:0010634) |
| 0.1 | 0.4 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.1 | 0.5 | GO:0035678 | neuromast hair cell morphogenesis(GO:0035678) |
| 0.1 | 8.3 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.1 | 1.0 | GO:0061099 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 0.9 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 0.5 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 1.3 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 0.8 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.6 | GO:0032196 | transposition(GO:0032196) |
| 0.1 | 3.2 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.1 | 0.2 | GO:1904103 | regulation of convergent extension involved in gastrulation(GO:1904103) |
| 0.1 | 2.2 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 0.5 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 1.5 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 1.0 | GO:0031269 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 2.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.4 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 2.6 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 0.6 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 0.9 | GO:1903039 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.1 | 8.7 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.1 | 2.9 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.1 | 0.6 | GO:0070601 | meiotic sister chromatid cohesion, centromeric(GO:0051754) centromeric sister chromatid cohesion(GO:0070601) |
| 0.1 | 0.6 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 1.0 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 3.8 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.1 | 1.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.9 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 2.1 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 0.6 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.4 | GO:0042987 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.1 | 2.0 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 0.8 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.4 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.8 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.1 | 1.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 1.3 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.1 | 6.6 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.1 | 1.0 | GO:0051122 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.8 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.1 | 4.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 1.9 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 0.8 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.2 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.6 | GO:0071684 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 4.9 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.1 | 2.3 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.1 | 0.7 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.1 | 0.3 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.1 | 0.6 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 4.6 | GO:0048738 | cardiac muscle tissue development(GO:0048738) |
| 0.1 | 2.3 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 1.0 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 2.4 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.1 | 0.5 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 1.5 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 2.9 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.1 | 0.3 | GO:0048025 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 2.5 | GO:0045103 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.1 | 0.7 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.4 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.1 | 3.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 1.7 | GO:0043542 | endothelial cell migration(GO:0043542) |
| 0.1 | 2.6 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 1.3 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.1 | 0.3 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.1 | 3.0 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.1 | 1.7 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 1.2 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.1 | 1.2 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 2.4 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.1 | 0.4 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.6 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 1.1 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 0.3 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 | 0.5 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.1 | 1.6 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.1 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 4.1 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.1 | 1.7 | GO:0021575 | hindbrain morphogenesis(GO:0021575) |
| 0.1 | 5.4 | GO:0060047 | heart contraction(GO:0060047) |
| 0.1 | 12.9 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.1 | 4.5 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.1 | 1.3 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.1 | 0.9 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.2 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.1 | 1.5 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 0.2 | GO:0015990 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.1 | 0.8 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 0.8 | GO:0030534 | adult behavior(GO:0030534) |
| 0.1 | 3.8 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.2 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 2.4 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.8 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.6 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.4 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.9 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 1.2 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 1.3 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.8 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.7 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 1.2 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 3.0 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 1.1 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 1.6 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.1 | GO:0021703 | locus ceruleus development(GO:0021703) |
| 0.0 | 0.4 | GO:0008585 | female gonad development(GO:0008585) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.5 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.5 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.8 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.3 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.8 | GO:0039021 | pronephric glomerulus development(GO:0039021) |
| 0.0 | 0.4 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.4 | GO:0030212 | hyaluronan metabolic process(GO:0030212) hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.3 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) sensory perception of taste(GO:0050909) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 3.5 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 0.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.2 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.9 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 16.3 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.4 | GO:0097581 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.0 | 0.3 | GO:0019320 | hexose catabolic process(GO:0019320) |
| 0.0 | 0.7 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.4 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 1.0 | GO:0008589 | regulation of smoothened signaling pathway(GO:0008589) |
| 0.0 | 1.0 | GO:1901342 | regulation of vasculature development(GO:1901342) |
| 0.0 | 0.8 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.2 | GO:0030819 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 1.1 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.0 | 0.6 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.4 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.2 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.2 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.0 | 0.5 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.5 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.0 | 0.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 1.1 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.2 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 1.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.7 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.6 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 1.5 | GO:0051604 | protein maturation(GO:0051604) |
| 0.0 | 0.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.6 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.2 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 1.3 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 1.6 | GO:0051216 | cartilage development(GO:0051216) |
| 0.0 | 0.5 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.2 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) positive regulation of dendrite development(GO:1900006) |
| 0.0 | 0.2 | GO:0048634 | regulation of muscle organ development(GO:0048634) |
| 0.0 | 0.7 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.1 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.7 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 1.1 | GO:0007051 | spindle organization(GO:0007051) |
| 0.0 | 1.0 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 0.1 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.9 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.9 | 19.8 | GO:0031430 | M band(GO:0031430) |
| 0.9 | 9.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.8 | 3.3 | GO:0031673 | H zone(GO:0031673) |
| 0.8 | 5.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.8 | 3.8 | GO:0008091 | spectrin(GO:0008091) |
| 0.7 | 4.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.6 | 1.9 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.6 | 4.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.6 | 3.5 | GO:0031672 | A band(GO:0031672) |
| 0.5 | 3.7 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.5 | 4.8 | GO:0030315 | T-tubule(GO:0030315) |
| 0.5 | 2.1 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.5 | 2.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.5 | 2.1 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.5 | 9.9 | GO:0016528 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.5 | 1.0 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.5 | 9.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.5 | 20.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.5 | 41.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.4 | 2.2 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.4 | 18.1 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.4 | 6.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.4 | 19.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.4 | 2.9 | GO:0046930 | pore complex(GO:0046930) |
| 0.4 | 1.4 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.4 | 5.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.3 | 2.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.3 | 4.8 | GO:0002102 | podosome(GO:0002102) |
| 0.3 | 1.0 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.3 | 3.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.3 | 2.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.3 | 28.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.3 | 0.9 | GO:1990879 | CST complex(GO:1990879) |
| 0.3 | 8.1 | GO:0008305 | integrin complex(GO:0008305) |
| 0.3 | 0.8 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.3 | 2.8 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.2 | 1.0 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.2 | 1.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.2 | 1.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 1.8 | GO:0070938 | contractile ring(GO:0070938) |
| 0.2 | 2.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 20.7 | GO:0030055 | cell-substrate junction(GO:0030055) |
| 0.2 | 3.0 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.2 | 1.5 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.2 | 4.7 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.2 | 0.6 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 13.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 0.8 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 0.6 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 0.7 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.1 | 1.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.8 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 0.8 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.5 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 1.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 1.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 1.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 2.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 2.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 1.9 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 6.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 4.8 | GO:0022626 | cytosolic ribosome(GO:0022626) cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.4 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 1.5 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.1 | 2.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 6.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 2.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 3.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 2.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.9 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 0.8 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 1.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.9 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 4.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 1.6 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 8.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 5.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 0.2 | GO:0070743 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.1 | 7.4 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 0.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 1.1 | GO:0098844 | postsynaptic endocytic zone(GO:0098843) postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 2.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.9 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 66.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 0.4 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.6 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.2 | GO:0097519 | DNA recombinase complex(GO:0097519) |
| 0.0 | 0.5 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 1.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 2.4 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 13.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 11.6 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 2.1 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 0.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 11.0 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.6 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.6 | GO:0000780 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 1.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 3.5 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.1 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.6 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 3.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.7 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.8 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.2 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.1 | GO:0035060 | brahma complex(GO:0035060) |
| 0.0 | 0.3 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.3 | GO:0005635 | nuclear envelope(GO:0005635) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 11.7 | GO:0071253 | connexin binding(GO:0071253) |
| 2.0 | 26.2 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 1.7 | 11.6 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 1.5 | 4.6 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 1.1 | 3.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 1.1 | 12.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 1.0 | 3.1 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 1.0 | 3.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.9 | 6.6 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.9 | 3.7 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.9 | 5.4 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.9 | 3.6 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.8 | 3.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.8 | 3.2 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.8 | 7.8 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.7 | 3.0 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.7 | 3.0 | GO:0031005 | filamin binding(GO:0031005) |
| 0.7 | 9.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.7 | 7.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.7 | 2.1 | GO:1990715 | mRNA CDS binding(GO:1990715) sequence-specific mRNA binding(GO:1990825) |
| 0.7 | 7.5 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.7 | 25.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.7 | 2.7 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.7 | 4.0 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.6 | 1.9 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.6 | 2.5 | GO:0102345 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.6 | 6.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.6 | 3.5 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.6 | 1.7 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.6 | 2.9 | GO:0015288 | porin activity(GO:0015288) |
| 0.5 | 1.5 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.4 | 1.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.4 | 19.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.4 | 7.5 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.4 | 7.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 2.0 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.3 | 3.5 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.3 | 8.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.3 | 1.5 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.3 | 2.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.3 | 2.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.3 | 0.8 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.3 | 0.8 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.3 | 5.6 | GO:0005344 | oxygen transporter activity(GO:0005344) oxygen binding(GO:0019825) |
| 0.3 | 15.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.3 | 2.6 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.3 | 0.8 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.2 | 2.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 1.7 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.2 | 0.7 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.2 | 3.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 2.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 3.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.2 | 2.6 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.2 | 1.2 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.2 | 8.0 | GO:0017022 | myosin binding(GO:0017022) |
| 0.2 | 4.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 3.7 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 2.6 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.2 | 2.8 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.2 | 2.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 2.9 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 3.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 1.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.2 | 3.8 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 0.8 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.2 | 0.6 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.2 | 2.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 0.9 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.2 | 0.9 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 2.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.2 | 3.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 2.1 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.2 | 1.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 1.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.2 | 2.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 0.8 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.2 | 4.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 0.6 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.2 | 1.8 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 1.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 2.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 4.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 1.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 1.4 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.1 | 0.4 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.1 | 18.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.4 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 1.8 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 0.8 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 2.0 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 1.0 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.1 | 1.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.6 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 0.6 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.1 | 0.7 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.4 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.1 | 2.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 2.5 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 1.0 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 3.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 10.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 38.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 1.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 4.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 4.9 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 0.4 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.1 | 1.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 4.2 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 4.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.8 | GO:0070696 | receptor serine/threonine kinase binding(GO:0033612) transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.1 | 1.0 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 2.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 1.7 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 1.1 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 0.6 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 6.6 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 8.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 1.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 1.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.8 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.4 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 4.9 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.1 | 6.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 1.7 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.5 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 2.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.8 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 0.3 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 1.9 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 1.8 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 3.2 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.1 | 0.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 4.3 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.1 | 1.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 2.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 1.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.4 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.6 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 0.5 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.1 | 1.0 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 2.4 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 1.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 1.0 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 29.9 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.1 | 37.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 0.2 | GO:0042164 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.1 | 8.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.8 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 0.4 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.1 | 0.2 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.1 | 0.4 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.1 | 0.8 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 0.8 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.1 | 0.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 2.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.0 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.1 | 0.6 | GO:0031420 | potassium ion binding(GO:0030955) alkali metal ion binding(GO:0031420) |
| 0.1 | 1.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 3.6 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 6.1 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 2.6 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.6 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.7 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.5 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 1.0 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.1 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.7 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 1.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.6 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.3 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 1.0 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.4 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.5 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.4 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 4.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.5 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.4 | GO:0034061 | DNA-directed DNA polymerase activity(GO:0003887) DNA polymerase activity(GO:0034061) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.7 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.8 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.6 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 1.4 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 0.4 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 8.7 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.7 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.0 | 0.3 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.0 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 1.4 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.8 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.5 | 12.0 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.3 | 7.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.3 | 4.3 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.3 | 11.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.3 | 10.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 5.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.2 | 1.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.2 | 5.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 9.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 3.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 3.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.2 | 4.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.2 | 3.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 26.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 2.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 3.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 3.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.2 | 3.0 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.2 | 2.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 4.5 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.2 | 4.2 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 2.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 2.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 6.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.5 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 1.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 0.8 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 13.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 12.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 0.9 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 1.1 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 1.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 0.7 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 1.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 0.5 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 0.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 3.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.5 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.5 | 13.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.4 | 5.8 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.4 | 4.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.4 | 5.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 15.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 4.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.2 | 1.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 6.2 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 6.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.2 | 2.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 4.0 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 1.7 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 3.8 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.2 | 0.9 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 2.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 6.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.2 | 3.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 0.9 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 2.5 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 5.8 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 2.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 2.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.8 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.1 | 1.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 4.0 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 1.0 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 1.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.9 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 3.7 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 1.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 5.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 0.6 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 3.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 2.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 8.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 1.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 1.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 2.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.0 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.1 | 0.8 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 0.9 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 0.6 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 3.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.7 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 2.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 0.3 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 3.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.9 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.1 | 0.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 0.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 1.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.6 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 1.2 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 5.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 3.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 1.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.6 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |