Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
Results for tcf7
Z-value: 0.64
Transcription factors associated with tcf7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tcf7
|
ENSDARG00000038672 | transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tcf7 | dr11_v1_chr21_+_45268112_45268112 | 0.21 | 4.0e-02 | Click! |
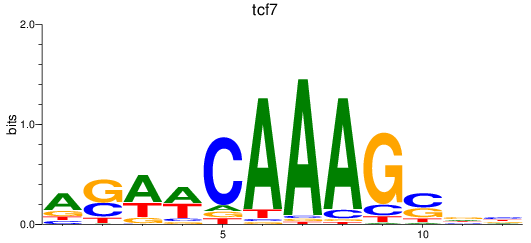
Activity profile of tcf7 motif
Sorted Z-values of tcf7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_5103349 | 5.85 |
ENSDART00000083474
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr21_+_20901505 | 5.31 |
ENSDART00000132741
|
c7b
|
complement component 7b |
| chr6_-_35487413 | 4.76 |
ENSDART00000102461
|
rgs8
|
regulator of G protein signaling 8 |
| chr18_+_17611627 | 4.05 |
ENSDART00000046891
|
cetp
|
cholesteryl ester transfer protein, plasma |
| chr6_-_35488180 | 3.56 |
ENSDART00000183258
|
rgs8
|
regulator of G protein signaling 8 |
| chr4_+_10366532 | 3.55 |
ENSDART00000189901
|
kcnd2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr7_+_38750871 | 3.41 |
ENSDART00000114238
ENSDART00000052325 ENSDART00000137001 |
f2
|
coagulation factor II (thrombin) |
| chr10_-_26744131 | 3.14 |
ENSDART00000020096
ENSDART00000162710 ENSDART00000179853 |
fgf13b
|
fibroblast growth factor 13b |
| chr9_-_9228941 | 3.05 |
ENSDART00000121665
|
cbsb
|
cystathionine-beta-synthase b |
| chr9_+_46745348 | 2.82 |
ENSDART00000025256
|
igfbp2b
|
insulin-like growth factor binding protein 2b |
| chr8_-_30979494 | 2.72 |
ENSDART00000138959
|
si:ch211-251j10.3
|
si:ch211-251j10.3 |
| chr10_+_21677058 | 2.59 |
ENSDART00000171499
ENSDART00000157516 |
pcdh1gb2
|
protocadherin 1 gamma b 2 |
| chr20_+_22666548 | 2.57 |
ENSDART00000147520
|
lnx1
|
ligand of numb-protein X 1 |
| chr10_+_21801030 | 2.54 |
ENSDART00000160881
ENSDART00000187969 |
pcdh1g30
|
protocadherin 1 gamma 30 |
| chr2_+_47623202 | 2.45 |
ENSDART00000154465
|
si:ch211-165b10.3
|
si:ch211-165b10.3 |
| chr20_+_30445971 | 2.43 |
ENSDART00000153150
|
myt1la
|
myelin transcription factor 1-like, a |
| chr1_-_38709551 | 2.41 |
ENSDART00000128794
|
gpm6ab
|
glycoprotein M6Ab |
| chr15_-_12545683 | 2.40 |
ENSDART00000162807
|
scn2b
|
sodium channel, voltage-gated, type II, beta |
| chr3_-_26190804 | 2.39 |
ENSDART00000136001
|
ypel3
|
yippee-like 3 |
| chr15_+_5901970 | 2.34 |
ENSDART00000114134
|
wrb
|
tryptophan rich basic protein |
| chr12_+_24561832 | 2.34 |
ENSDART00000088159
|
nrxn1a
|
neurexin 1a |
| chr16_+_23913943 | 2.32 |
ENSDART00000175404
ENSDART00000129525 |
apoa4b.1
|
apolipoprotein A-IV b, tandem duplicate 1 |
| chr8_+_26859639 | 2.25 |
ENSDART00000133440
|
prdm2a
|
PR domain containing 2, with ZNF domain a |
| chr14_-_2196267 | 2.18 |
ENSDART00000161674
ENSDART00000125674 |
pcdh2ab8
pcdh2ab9
|
protocadherin 2 alpha b 8 protocadherin 2 alpha b 9 |
| chr13_-_9895564 | 2.14 |
ENSDART00000169831
ENSDART00000142629 |
si:ch211-117n7.6
|
si:ch211-117n7.6 |
| chr2_-_44038698 | 2.07 |
ENSDART00000079582
ENSDART00000146804 |
kirrel1b
|
kirre like nephrin family adhesion molecule 1b |
| chr7_-_26308098 | 2.06 |
ENSDART00000146440
ENSDART00000146935 ENSDART00000164627 |
zgc:77439
|
zgc:77439 |
| chr17_+_3128273 | 2.03 |
ENSDART00000122453
|
zgc:136872
|
zgc:136872 |
| chr21_+_13387965 | 2.02 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr19_-_13774502 | 1.96 |
ENSDART00000159711
|
epb41a
|
erythrocyte membrane protein band 4.1a |
| chr8_-_1266181 | 1.90 |
ENSDART00000148654
ENSDART00000149924 |
cdc14b
|
cell division cycle 14B |
| chr5_-_28149767 | 1.90 |
ENSDART00000051515
|
zgc:110329
|
zgc:110329 |
| chr21_+_15709061 | 1.85 |
ENSDART00000065772
|
ddt
|
D-dopachrome tautomerase |
| chr11_-_23687158 | 1.83 |
ENSDART00000189599
|
pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr2_-_37043540 | 1.83 |
ENSDART00000131834
|
gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr16_+_25137483 | 1.77 |
ENSDART00000155666
|
znf576.1
|
zinc finger protein 576, tandem duplicate 1 |
| chr13_+_27073901 | 1.76 |
ENSDART00000146227
|
slc24a3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 |
| chr5_-_24231139 | 1.75 |
ENSDART00000143492
|
senp3a
|
SUMO1/sentrin/SMT3 specific peptidase 3a |
| chr15_+_6109861 | 1.73 |
ENSDART00000185154
|
PCP4 (1 of many)
|
Purkinje cell protein 4 |
| chr5_-_23696926 | 1.72 |
ENSDART00000021462
|
rnf128a
|
ring finger protein 128a |
| chr18_-_13360106 | 1.71 |
ENSDART00000091512
|
cmip
|
c-Maf inducing protein |
| chr24_-_5932982 | 1.70 |
ENSDART00000138412
ENSDART00000135124 ENSDART00000007373 |
acbd5a
|
acyl-CoA binding domain containing 5a |
| chr7_+_14632157 | 1.70 |
ENSDART00000161264
|
ntrk3b
|
neurotrophic tyrosine kinase, receptor, type 3b |
| chr1_-_26023678 | 1.69 |
ENSDART00000054202
|
si:ch211-145b13.5
|
si:ch211-145b13.5 |
| chr9_+_38292947 | 1.69 |
ENSDART00000146663
|
tfcp2l1
|
transcription factor CP2-like 1 |
| chr21_-_4032650 | 1.68 |
ENSDART00000151648
|
ntng2b
|
netrin g2b |
| chr4_-_23839789 | 1.67 |
ENSDART00000143571
|
usp6nl
|
USP6 N-terminal like |
| chr21_+_36774542 | 1.66 |
ENSDART00000130790
|
prr7
|
proline rich 7 (synaptic) |
| chr14_-_2358774 | 1.62 |
ENSDART00000164809
|
si:ch73-233f7.5
|
si:ch73-233f7.5 |
| chr14_-_2004291 | 1.61 |
ENSDART00000114039
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr22_-_20126230 | 1.61 |
ENSDART00000138688
|
creb3l3a
|
cAMP responsive element binding protein 3-like 3a |
| chr12_+_27068525 | 1.60 |
ENSDART00000188634
|
srcap
|
Snf2-related CREBBP activator protein |
| chr24_+_5840258 | 1.55 |
ENSDART00000087034
|
trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr25_-_12788370 | 1.50 |
ENSDART00000158551
|
slc7a5
|
solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
| chr9_+_33009284 | 1.49 |
ENSDART00000036926
|
vangl1
|
VANGL planar cell polarity protein 1 |
| chr16_-_12097394 | 1.44 |
ENSDART00000103944
|
pex5
|
peroxisomal biogenesis factor 5 |
| chr17_+_24597001 | 1.42 |
ENSDART00000191834
|
rlf
|
rearranged L-myc fusion |
| chr19_-_7033636 | 1.40 |
ENSDART00000122815
ENSDART00000123443 ENSDART00000124094 |
daxx
|
death-domain associated protein |
| chr17_+_3993772 | 1.40 |
ENSDART00000170822
ENSDART00000168613 ENSDART00000186174 |
tmx4
|
thioredoxin-related transmembrane protein 4 |
| chr4_-_2525916 | 1.39 |
ENSDART00000134123
ENSDART00000132581 ENSDART00000019508 |
csrp2
|
cysteine and glycine-rich protein 2 |
| chr10_+_19569052 | 1.39 |
ENSDART00000058425
|
CABZ01059627.1
|
|
| chr5_+_60590796 | 1.38 |
ENSDART00000159859
|
tmem132e
|
transmembrane protein 132E |
| chr8_+_17869225 | 1.38 |
ENSDART00000080079
|
slc44a5b
|
solute carrier family 44, member 5b |
| chr15_-_19677511 | 1.37 |
ENSDART00000043743
|
si:dkey-4p15.3
|
si:dkey-4p15.3 |
| chr1_-_41374117 | 1.36 |
ENSDART00000074777
|
htt
|
huntingtin |
| chr4_+_16827390 | 1.34 |
ENSDART00000113344
|
kiss2
|
kisspeptin 2 |
| chr4_+_25651720 | 1.33 |
ENSDART00000100693
ENSDART00000100717 |
acot16
|
acyl-CoA thioesterase 16 |
| chr4_-_14531687 | 1.27 |
ENSDART00000182093
ENSDART00000159447 |
plxnb2a
|
plexin b2a |
| chr1_-_50293946 | 1.27 |
ENSDART00000053028
ENSDART00000125099 |
tbck
|
TBC1 domain containing kinase |
| chr9_+_13733468 | 1.26 |
ENSDART00000165954
ENSDART00000160355 ENSDART00000081040 ENSDART00000138254 ENSDART00000081015 ENSDART00000141314 |
abi2a
|
abl-interactor 2a |
| chr7_-_34329527 | 1.25 |
ENSDART00000173454
|
madd
|
MAP-kinase activating death domain |
| chr10_+_45128375 | 1.24 |
ENSDART00000164805
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr11_-_42099645 | 1.23 |
ENSDART00000173312
|
abhd6a
|
abhydrolase domain containing 6a |
| chr17_+_47116500 | 1.22 |
ENSDART00000186627
|
CLIP4
|
si:dkeyp-47f9.4 |
| chr15_-_23761580 | 1.21 |
ENSDART00000137918
|
bbc3
|
BCL2 binding component 3 |
| chr17_+_11500628 | 1.21 |
ENSDART00000155660
|
efcab2
|
EF-hand calcium binding domain 2 |
| chr5_+_42124706 | 1.18 |
ENSDART00000020044
ENSDART00000156372 |
shpk
|
sedoheptulokinase |
| chr5_+_20453874 | 1.18 |
ENSDART00000124545
ENSDART00000008402 |
sart3
|
squamous cell carcinoma antigen recognized by T cells 3 |
| chr24_-_6546479 | 1.17 |
ENSDART00000160538
|
arhgap21a
|
Rho GTPase activating protein 21a |
| chr23_-_27061072 | 1.14 |
ENSDART00000124482
|
zgc:66440
|
zgc:66440 |
| chr17_-_20897250 | 1.13 |
ENSDART00000088106
|
ank3b
|
ankyrin 3b |
| chr24_-_41267184 | 1.12 |
ENSDART00000063504
|
xylb
|
xylulokinase homolog (H. influenzae) |
| chr11_+_44226200 | 1.11 |
ENSDART00000191417
|
gnb4b
|
guanine nucleotide binding protein (G protein), beta polypeptide 4b |
| chr5_+_43470544 | 1.11 |
ENSDART00000111587
|
rhobtb2a
|
Rho-related BTB domain containing 2a |
| chr6_+_47843760 | 1.10 |
ENSDART00000140943
|
padi2
|
peptidyl arginine deiminase, type II |
| chr6_-_57938043 | 1.10 |
ENSDART00000171073
|
tox2
|
TOX high mobility group box family member 2 |
| chr24_-_6345647 | 1.10 |
ENSDART00000108994
|
zgc:174877
|
zgc:174877 |
| chr3_+_19914332 | 1.07 |
ENSDART00000078982
|
vat1
|
vesicle amine transport 1 |
| chr25_+_18964782 | 1.06 |
ENSDART00000017299
|
tdg.1
|
thymine DNA glycosylase, tandem duplicate 1 |
| chr5_-_16996482 | 1.06 |
ENSDART00000144501
|
galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr14_-_2036604 | 1.05 |
ENSDART00000192446
|
BX005294.2
|
|
| chr15_-_17190540 | 1.03 |
ENSDART00000154801
|
itgae.2
|
integrin, alpha E, tandem duplicate 2 |
| chr5_-_56964547 | 1.03 |
ENSDART00000074400
|
tia1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr3_+_15773991 | 1.03 |
ENSDART00000089923
|
znf652
|
zinc finger protein 652 |
| chr20_-_11178022 | 1.01 |
ENSDART00000152246
|
flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr17_+_32500387 | 0.99 |
ENSDART00000018423
|
ywhaqb
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide b |
| chr20_+_23315242 | 0.96 |
ENSDART00000132248
|
fryl
|
furry homolog, like |
| chr16_-_41131578 | 0.95 |
ENSDART00000102649
ENSDART00000145956 |
ptpn23a
|
protein tyrosine phosphatase, non-receptor type 23, a |
| chr14_-_36863432 | 0.95 |
ENSDART00000158052
|
rnf130
|
ring finger protein 130 |
| chr21_-_40782393 | 0.94 |
ENSDART00000075808
|
apbb3
|
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr24_+_26017094 | 0.93 |
ENSDART00000137851
|
tfr1b
|
transferrin receptor 1b |
| chr4_-_2036620 | 0.92 |
ENSDART00000150490
|
si:dkey-97m3.1
|
si:dkey-97m3.1 |
| chr4_-_1908179 | 0.90 |
ENSDART00000139586
|
ano6
|
anoctamin 6 |
| chr10_+_4046448 | 0.90 |
ENSDART00000123086
ENSDART00000052268 |
pitpnb
|
phosphatidylinositol transfer protein, beta |
| chr13_+_28732101 | 0.90 |
ENSDART00000015773
|
ldb1a
|
LIM domain binding 1a |
| chr2_-_59376735 | 0.88 |
ENSDART00000193624
|
ftr38
|
finTRIM family, member 38 |
| chr12_+_48204891 | 0.86 |
ENSDART00000190534
ENSDART00000164427 |
ndr2
|
nodal-related 2 |
| chr24_-_11908115 | 0.86 |
ENSDART00000184329
|
tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr2_-_55903520 | 0.85 |
ENSDART00000128828
|
cplx4c
|
complexin 4c |
| chr23_+_1702624 | 0.85 |
ENSDART00000149357
|
rabggta
|
Rab geranylgeranyltransferase, alpha subunit |
| chr21_-_18275226 | 0.82 |
ENSDART00000126672
ENSDART00000135239 |
brd3a
|
bromodomain containing 3a |
| chr18_+_18861359 | 0.82 |
ENSDART00000144605
|
pllp
|
plasmolipin |
| chr4_+_47174366 | 0.80 |
ENSDART00000170732
|
si:dkey-26m3.1
|
si:dkey-26m3.1 |
| chr21_-_37889727 | 0.78 |
ENSDART00000163612
ENSDART00000180958 |
gabrb4
|
gamma-aminobutyric acid (GABA) A receptor, beta 4 |
| chr7_+_39688208 | 0.78 |
ENSDART00000189682
|
tbc1d14
|
TBC1 domain family, member 14 |
| chr14_+_24277556 | 0.76 |
ENSDART00000122660
|
hnrnpa0a
|
heterogeneous nuclear ribonucleoprotein A0a |
| chr2_+_37975026 | 0.76 |
ENSDART00000034802
|
si:rp71-1g18.13
|
si:rp71-1g18.13 |
| chr8_-_2153147 | 0.74 |
ENSDART00000124093
|
si:dkeyp-117b11.1
|
si:dkeyp-117b11.1 |
| chr18_+_38775277 | 0.73 |
ENSDART00000186129
|
fam214a
|
family with sequence similarity 214, member A |
| chr22_+_8753092 | 0.72 |
ENSDART00000140720
|
si:dkey-182g1.2
|
si:dkey-182g1.2 |
| chr16_+_1353894 | 0.72 |
ENSDART00000148426
|
celf3b
|
cugbp, Elav-like family member 3b |
| chr1_-_54706039 | 0.72 |
ENSDART00000083633
|
exosc1
|
exosome component 1 |
| chr18_-_6534516 | 0.71 |
ENSDART00000009217
|
ddx11
|
DEAD/H (Asp-Glu-Ala-Asp/His) box helicase 11 |
| chr4_-_30422325 | 0.70 |
ENSDART00000158444
|
znf1114
|
zinc finger protein 1114 |
| chr3_-_1146497 | 0.68 |
ENSDART00000149061
|
si:ch73-211l13.2
|
si:ch73-211l13.2 |
| chr13_+_33268657 | 0.67 |
ENSDART00000002095
|
tmem39b
|
transmembrane protein 39B |
| chr14_+_22293787 | 0.65 |
ENSDART00000157967
|
atp10b
|
ATPase phospholipid transporting 10B |
| chr5_+_44064764 | 0.65 |
ENSDART00000143843
|
si:dkey-84j12.1
|
si:dkey-84j12.1 |
| chr15_+_23784842 | 0.63 |
ENSDART00000192889
ENSDART00000138375 |
ift20
|
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr10_-_11261565 | 0.63 |
ENSDART00000146727
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr10_-_21542702 | 0.61 |
ENSDART00000146761
ENSDART00000134502 |
zgc:165539
|
zgc:165539 |
| chr6_-_29105727 | 0.61 |
ENSDART00000184355
|
fam69ab
|
family with sequence similarity 69, member Ab |
| chr21_+_22423286 | 0.60 |
ENSDART00000133190
|
capslb
|
calcyphosine-like b |
| chr14_-_2193248 | 0.59 |
ENSDART00000128659
|
pcdh2ab10
|
protocadherin 2 alpha b 10 |
| chr7_-_29223614 | 0.57 |
ENSDART00000173598
|
herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr22_+_12161738 | 0.57 |
ENSDART00000168857
|
ccnt2b
|
cyclin T2b |
| chr17_+_13664442 | 0.57 |
ENSDART00000171689
|
lrfn5a
|
leucine rich repeat and fibronectin type III domain containing 5a |
| chr11_-_17755444 | 0.54 |
ENSDART00000154627
|
eogt
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr12_+_46512881 | 0.54 |
ENSDART00000105454
|
CU855711.1
|
|
| chr17_+_43595692 | 0.54 |
ENSDART00000156271
|
cfap99
|
cilia and flagella associated protein 99 |
| chr2_-_22152797 | 0.51 |
ENSDART00000145188
|
cyp7a1
|
cytochrome P450, family 7, subfamily A, polypeptide 1 |
| chr6_-_51101834 | 0.49 |
ENSDART00000092493
|
ptprt
|
protein tyrosine phosphatase, receptor type, t |
| chr23_+_27051919 | 0.48 |
ENSDART00000054237
|
ptges3a
|
prostaglandin E synthase 3a (cytosolic) |
| chr16_-_12097558 | 0.47 |
ENSDART00000123142
|
pex5
|
peroxisomal biogenesis factor 5 |
| chr9_-_1604601 | 0.45 |
ENSDART00000143130
|
agps
|
alkylglycerone phosphate synthase |
| chr1_-_16665044 | 0.44 |
ENSDART00000040434
|
asah1b
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1b |
| chr25_-_20574859 | 0.43 |
ENSDART00000067288
|
adal
|
adenosine deaminase-like |
| chr1_+_32341484 | 0.43 |
ENSDART00000137156
|
pudp
|
pseudouridine 5'-phosphatase |
| chr4_+_69375564 | 0.42 |
ENSDART00000150334
|
si:dkey-246j6.1
|
si:dkey-246j6.1 |
| chr14_-_36862745 | 0.42 |
ENSDART00000109293
|
rnf130
|
ring finger protein 130 |
| chr17_-_6076084 | 0.39 |
ENSDART00000058890
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr14_-_6943934 | 0.39 |
ENSDART00000126279
|
clk4a
|
CDC-like kinase 4a |
| chr25_+_3294150 | 0.37 |
ENSDART00000030683
|
tmpob
|
thymopoietin b |
| chr14_-_38827442 | 0.37 |
ENSDART00000160000
|
spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr12_+_46462090 | 0.36 |
ENSDART00000130748
|
BX005305.2
|
|
| chr5_+_67371650 | 0.36 |
ENSDART00000142156
|
sebox
|
SEBOX homeobox |
| chr3_+_5297493 | 0.35 |
ENSDART00000138596
|
si:ch211-150d5.3
|
si:ch211-150d5.3 |
| chr24_+_11105786 | 0.34 |
ENSDART00000175182
|
prlh2
|
prolactin releasing hormone 2 |
| chr2_-_16254782 | 0.34 |
ENSDART00000133708
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr17_-_20897407 | 0.34 |
ENSDART00000149481
|
ank3b
|
ankyrin 3b |
| chr10_+_23099365 | 0.31 |
ENSDART00000022170
|
LTN1
|
si:dkey-175g6.5 |
| chr19_-_18135724 | 0.30 |
ENSDART00000186609
|
cbx3a
|
chromobox homolog 3a (HP1 gamma homolog, Drosophila) |
| chr22_+_21549419 | 0.30 |
ENSDART00000139411
|
plpp2b
|
phospholipid phosphatase 2b |
| chr7_-_54677143 | 0.29 |
ENSDART00000163748
|
ccnd1
|
cyclin D1 |
| chr21_-_22474362 | 0.29 |
ENSDART00000169659
|
myo5b
|
myosin VB |
| chr15_-_47822597 | 0.29 |
ENSDART00000193236
ENSDART00000161391 |
CZQB01095947.1
|
|
| chr7_+_35075847 | 0.28 |
ENSDART00000193469
ENSDART00000037346 |
ctrb1
|
chymotrypsinogen B1 |
| chr10_-_11261386 | 0.26 |
ENSDART00000189946
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr19_+_42469058 | 0.24 |
ENSDART00000076915
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr7_+_36898622 | 0.22 |
ENSDART00000190773
|
tox3
|
TOX high mobility group box family member 3 |
| chr17_+_14711765 | 0.22 |
ENSDART00000012889
|
cx28.6
|
connexin 28.6 |
| chr7_-_57509447 | 0.21 |
ENSDART00000051973
ENSDART00000147036 |
sirt3
|
sirtuin 3 |
| chr11_-_3381343 | 0.20 |
ENSDART00000002545
|
mcrs1
|
microspherule protein 1 |
| chr3_+_41558682 | 0.20 |
ENSDART00000157023
|
card11
|
caspase recruitment domain family, member 11 |
| chr9_-_28616436 | 0.18 |
ENSDART00000136985
|
si:ch73-7i4.2
|
si:ch73-7i4.2 |
| chr4_-_32192868 | 0.18 |
ENSDART00000164675
|
CR450785.2
|
|
| chr11_-_3380959 | 0.18 |
ENSDART00000187157
|
mcrs1
|
microspherule protein 1 |
| chr5_+_32791245 | 0.17 |
ENSDART00000077189
|
ier5l
|
immediate early response 5-like |
| chr17_-_20695998 | 0.17 |
ENSDART00000063613
|
ccdc6a
|
coiled-coil domain containing 6a |
| chr1_-_50438247 | 0.17 |
ENSDART00000114098
|
dkk2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr2_-_43739740 | 0.15 |
ENSDART00000113849
|
kif5ba
|
kinesin family member 5B, a |
| chr1_+_24376394 | 0.14 |
ENSDART00000102542
|
zgc:171517
|
zgc:171517 |
| chr1_-_17675037 | 0.13 |
ENSDART00000142689
|
si:dkey-256e7.5
|
si:dkey-256e7.5 |
| chr16_+_7703693 | 0.12 |
ENSDART00000125111
ENSDART00000149769 ENSDART00000149752 ENSDART00000081407 |
cnot10
|
CCR4-NOT transcription complex, subunit 10 |
| chr19_+_43037657 | 0.11 |
ENSDART00000168263
ENSDART00000184771 ENSDART00000164453 ENSDART00000165202 |
pum1
|
pumilio RNA-binding family member 1 |
| chr20_-_4031475 | 0.11 |
ENSDART00000112053
|
fam89a
|
family with sequence similarity 89, member A |
| chr4_-_3064101 | 0.10 |
ENSDART00000135701
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr17_-_6076266 | 0.10 |
ENSDART00000171084
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr14_+_989733 | 0.10 |
ENSDART00000161487
ENSDART00000127317 |
si:ch73-308l14.2
|
si:ch73-308l14.2 |
| chr23_-_7826849 | 0.09 |
ENSDART00000157612
|
myt1b
|
myelin transcription factor 1b |
| chr15_-_1484795 | 0.08 |
ENSDART00000129356
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr9_-_37613792 | 0.07 |
ENSDART00000138345
|
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
| chr23_-_19977214 | 0.07 |
ENSDART00000054659
|
ccnq
|
cyclin Q |
| chr4_-_77114795 | 0.04 |
ENSDART00000144849
|
CU467646.2
|
|
| chr14_+_1240604 | 0.04 |
ENSDART00000188258
|
adad1
|
adenosine deaminase domain containing 1 (testis-specific) |
| chr9_-_12401871 | 0.02 |
ENSDART00000191901
|
nup35
|
nucleoporin 35 |
| chr14_+_1240419 | 0.00 |
ENSDART00000181248
|
adad1
|
adenosine deaminase domain containing 1 (testis-specific) |
Network of associatons between targets according to the STRING database.
First level regulatory network of tcf7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.3 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 1.4 | 4.1 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 1.0 | 3.0 | GO:0050666 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.5 | 5.9 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.5 | 3.4 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.4 | 2.1 | GO:0052651 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.4 | 1.2 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.4 | 1.1 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.3 | 0.9 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.3 | 1.7 | GO:0030242 | pexophagy(GO:0030242) |
| 0.3 | 2.8 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.2 | 1.2 | GO:0090199 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.2 | 0.9 | GO:0033572 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.2 | 3.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.2 | 1.1 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.2 | 1.4 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.2 | 1.9 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.2 | 1.9 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.2 | 5.3 | GO:0019835 | cytolysis(GO:0019835) |
| 0.2 | 0.5 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.2 | 0.6 | GO:0035790 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.2 | 0.5 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.4 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 1.4 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.1 | 0.8 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 1.7 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.9 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.1 | 1.8 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 2.2 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 1.6 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 1.7 | GO:1990090 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.1 | 1.1 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 2.1 | GO:0039022 | pronephric duct development(GO:0039022) |
| 0.1 | 0.5 | GO:0008206 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.1 | 0.4 | GO:0046102 | adenosine catabolic process(GO:0006154) inosine metabolic process(GO:0046102) inosine biosynthetic process(GO:0046103) |
| 0.1 | 1.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.3 | GO:0003245 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 1.6 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 1.7 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 0.3 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 0.8 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 11.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.8 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 1.7 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 1.6 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 1.2 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.5 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 1.1 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.9 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 2.0 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 1.5 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.2 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.0 | 2.9 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.9 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.8 | GO:0048546 | digestive tract morphogenesis(GO:0048546) |
| 0.0 | 1.4 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.6 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 1.3 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.5 | GO:0060841 | venous blood vessel development(GO:0060841) |
| 0.0 | 2.1 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 3.1 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.0 | 0.7 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 2.1 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.8 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.5 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.1 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 1.9 | GO:0072659 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 1.0 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.1 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.9 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.3 | 5.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.3 | 4.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 0.8 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.2 | 2.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 1.6 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 2.1 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 2.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 3.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 4.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 1.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.7 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 2.8 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 0.7 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 0.6 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 1.0 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.8 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.6 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 1.0 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.9 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.8 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.0 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 1.8 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.8 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.9 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 3.1 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.0 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.6 | 3.5 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.5 | 5.9 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.3 | 1.7 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.3 | 2.8 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 1.9 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.2 | 0.9 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.2 | 2.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 0.8 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.2 | 5.0 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.2 | 1.6 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 1.8 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 2.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 0.5 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 1.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.9 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.9 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.9 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 1.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.5 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 1.1 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.1 | 0.4 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.1 | 1.7 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 1.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 1.8 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.4 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 0.5 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 1.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.3 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 1.1 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 1.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 1.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.7 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 0.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 1.4 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 1.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.6 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0070404 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.0 | 12.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.5 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 1.5 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 1.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 1.9 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 2.0 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 3.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.5 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.5 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 3.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.4 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 2.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.7 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.7 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.3 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 4.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 2.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 3.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 9.3 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 0.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 1.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 0.7 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 0.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 0.9 | REACTOME PI METABOLISM | Genes involved in PI Metabolism |
| 0.0 | 1.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |