Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
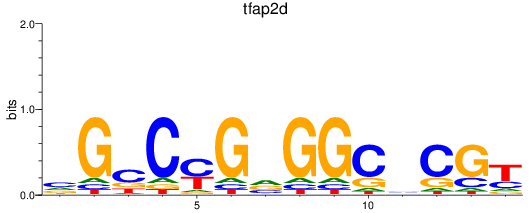
Results for tfap2d
Z-value: 0.42
Transcription factors associated with tfap2d
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tfap2d
|
ENSDARG00000023272 | transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tfap2d | dr11_v1_chr20_-_47731768_47731768 | -0.74 | 1.2e-17 | Click! |
Activity profile of tfap2d motif
Sorted Z-values of tfap2d motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_188102 | 7.36 |
ENSDART00000125391
ENSDART00000170463 |
CABZ01052268.1
|
|
| chr10_-_322769 | 5.09 |
ENSDART00000165244
|
akt2l
|
v-akt murine thymoma viral oncogene homolog 2, like |
| chr14_-_21064199 | 4.56 |
ENSDART00000172099
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr21_+_45685757 | 4.55 |
ENSDART00000160530
|
sec24a
|
SEC24 homolog A, COPII coat complex component |
| chr14_+_52369262 | 3.83 |
ENSDART00000169352
ENSDART00000157833 |
igfbp7
|
insulin-like growth factor binding protein 7 |
| chr4_-_77557279 | 3.44 |
ENSDART00000180113
|
AL935186.10
|
|
| chr17_-_868004 | 3.20 |
ENSDART00000112803
|
wdr20a
|
WD repeat domain 20a |
| chr16_+_54588930 | 2.94 |
ENSDART00000159174
|
dennd4b
|
DENN/MADD domain containing 4B |
| chr10_-_373575 | 2.81 |
ENSDART00000114487
|
DMPK
|
DM1 protein kinase |
| chr5_-_26936861 | 1.36 |
ENSDART00000191032
|
htra4
|
HtrA serine peptidase 4 |
| chr25_+_2415131 | 1.35 |
ENSDART00000162210
|
zmp:0000000932
|
zmp:0000000932 |
| chr6_+_27868146 | 1.19 |
ENSDART00000186620
|
rab6ba
|
RAB6B, member RAS oncogene family a |
| chr3_+_31680592 | 1.02 |
ENSDART00000172456
|
mylk5
|
myosin, light chain kinase 5 |
| chr23_-_41762797 | 0.97 |
ENSDART00000186564
|
stk35
|
serine/threonine kinase 35 |
| chr5_+_6796291 | 0.77 |
ENSDART00000166868
ENSDART00000165308 |
me2
|
malic enzyme 2, NAD(+)-dependent, mitochondrial |
| chr17_+_52823015 | 0.73 |
ENSDART00000160507
ENSDART00000186979 |
meis2a
|
Meis homeobox 2a |
| chr20_-_14925281 | 0.59 |
ENSDART00000152641
|
dnm3a
|
dynamin 3a |
| chr5_+_58520266 | 0.58 |
ENSDART00000108889
|
oafb
|
OAF homolog b (Drosophila) |
| chr13_-_36579086 | 0.50 |
ENSDART00000146671
|
lgals3a
|
lectin, galactoside binding soluble 3a |
| chr12_-_19007834 | 0.39 |
ENSDART00000153248
|
chadlb
|
chondroadherin-like b |
| chr1_-_23294753 | 0.38 |
ENSDART00000013263
|
ugdh
|
UDP-glucose 6-dehydrogenase |
Network of associatons between targets according to the STRING database.
First level regulatory network of tfap2d
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.6 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.3 | 4.5 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 2.9 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 3.8 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.1 | 0.5 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 0.8 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.4 | GO:0003188 | heart valve formation(GO:0003188) atrioventricular valve formation(GO:0003190) |
| 0.0 | 1.2 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.6 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 5.1 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.6 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.2 | 5.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.5 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.1 | 0.4 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.1 | 1.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 4.5 | GO:0000149 | SNARE binding(GO:0000149) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |