Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
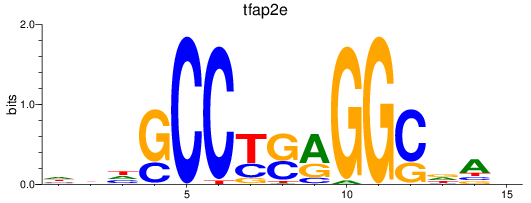
Results for tfap2e
Z-value: 0.58
Transcription factors associated with tfap2e
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tfap2e
|
ENSDARG00000008861 | transcription factor AP-2 epsilon |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tfap2e | dr11_v1_chr19_-_47456787_47456787 | 0.69 | 1.4e-14 | Click! |
Activity profile of tfap2e motif
Sorted Z-values of tfap2e motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_9472893 | 15.13 |
ENSDART00000045565
ENSDART00000137505 |
vamp1
|
vesicle-associated membrane protein 1 |
| chr15_-_16098531 | 12.97 |
ENSDART00000080377
|
aldoca
|
aldolase C, fructose-bisphosphate, a |
| chr15_-_44512461 | 7.80 |
ENSDART00000155456
|
gria4a
|
glutamate receptor, ionotropic, AMPA 4a |
| chr23_+_37323962 | 7.02 |
ENSDART00000102881
|
fam43b
|
family with sequence similarity 43, member B |
| chr2_+_24199276 | 6.76 |
ENSDART00000140575
|
map4l
|
microtubule associated protein 4 like |
| chr16_+_45571956 | 5.42 |
ENSDART00000143867
|
syngap1b
|
synaptic Ras GTPase activating protein 1b |
| chr24_-_6898302 | 5.02 |
ENSDART00000158646
|
dpp6a
|
dipeptidyl-peptidase 6a |
| chr6_-_10320676 | 4.78 |
ENSDART00000151247
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr19_-_10395683 | 4.41 |
ENSDART00000109488
|
zgc:194578
|
zgc:194578 |
| chr7_-_51476276 | 4.39 |
ENSDART00000082464
|
nhsl2
|
NHS-like 2 |
| chr2_+_27010439 | 4.37 |
ENSDART00000030547
|
cdh7a
|
cadherin 7a |
| chr14_+_29609245 | 4.35 |
ENSDART00000043058
|
TENM2
|
si:dkey-34l15.2 |
| chr23_+_2760573 | 4.33 |
ENSDART00000129719
|
top1
|
DNA topoisomerase I |
| chr16_-_29458806 | 4.28 |
ENSDART00000047931
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr9_+_32978302 | 4.17 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| chr21_-_21089781 | 4.16 |
ENSDART00000144361
|
ank1b
|
ankyrin 1, erythrocytic b |
| chr11_+_30253968 | 4.12 |
ENSDART00000157272
ENSDART00000003475 |
ppef1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr2_-_15041846 | 4.01 |
ENSDART00000139050
|
si:dkey-10f21.4
|
si:dkey-10f21.4 |
| chr19_+_32979331 | 3.92 |
ENSDART00000078066
|
spire1a
|
spire-type actin nucleation factor 1a |
| chr22_+_1440702 | 3.91 |
ENSDART00000165677
|
si:dkeyp-53d3.3
|
si:dkeyp-53d3.3 |
| chr16_-_46393154 | 3.84 |
ENSDART00000132154
|
si:ch73-59c19.1
|
si:ch73-59c19.1 |
| chr10_-_2526526 | 3.72 |
ENSDART00000191726
ENSDART00000192767 |
CU856539.1
|
|
| chr19_+_47311869 | 3.65 |
ENSDART00000136647
|
ext1c
|
exostoses (multiple) 1c |
| chr9_+_32358514 | 3.39 |
ENSDART00000144608
|
plcl1
|
phospholipase C like 1 |
| chr2_+_24199073 | 3.18 |
ENSDART00000144110
|
map4l
|
microtubule associated protein 4 like |
| chr16_-_17162485 | 3.14 |
ENSDART00000123011
|
iffo1b
|
intermediate filament family orphan 1b |
| chr16_+_36906693 | 3.08 |
ENSDART00000160645
|
si:ch73-215d9.1
|
si:ch73-215d9.1 |
| chr12_+_19036380 | 3.07 |
ENSDART00000153086
ENSDART00000181060 |
kctd17
|
potassium channel tetramerization domain containing 17 |
| chr19_+_1184878 | 3.01 |
ENSDART00000163539
|
scrt1a
|
scratch family zinc finger 1a |
| chr4_+_26972862 | 2.92 |
ENSDART00000011519
ENSDART00000138830 |
slc6a1l
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 1, like |
| chr7_-_23563092 | 2.91 |
ENSDART00000132275
|
gpr185b
|
G protein-coupled receptor 185 b |
| chr18_+_34478959 | 2.88 |
ENSDART00000059394
|
kcnab1a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 a |
| chr14_-_31465905 | 2.76 |
ENSDART00000173108
|
gpc3
|
glypican 3 |
| chr23_-_18668836 | 2.67 |
ENSDART00000138792
ENSDART00000051182 |
arhgap4b
|
Rho GTPase activating protein 4b |
| chr10_+_36029537 | 2.45 |
ENSDART00000165386
|
hmgb1a
|
high mobility group box 1a |
| chr5_+_63302660 | 2.27 |
ENSDART00000142131
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr21_-_39577517 | 2.19 |
ENSDART00000143566
|
srsf1b
|
serine/arginine-rich splicing factor 1b |
| chr23_-_9965148 | 2.07 |
ENSDART00000137308
|
plxnb1a
|
plexin b1a |
| chr3_-_54607166 | 2.04 |
ENSDART00000021977
|
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr10_+_5135842 | 1.99 |
ENSDART00000132627
ENSDART00000162434 |
zgc:113274
|
zgc:113274 |
| chr13_+_16342926 | 1.97 |
ENSDART00000105496
ENSDART00000124710 |
dlg5a
|
discs, large homolog 5a (Drosophila) |
| chr19_-_27462720 | 1.93 |
ENSDART00000135901
|
si:ch73-25f10.6
|
si:ch73-25f10.6 |
| chr7_+_39011355 | 1.61 |
ENSDART00000173855
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr22_-_57177 | 1.58 |
ENSDART00000163959
|
CABZ01085139.1
|
|
| chr7_+_39011852 | 1.56 |
ENSDART00000093009
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr15_+_44929497 | 1.53 |
ENSDART00000111373
|
pdgfd
|
platelet derived growth factor d |
| chr11_+_31323746 | 1.44 |
ENSDART00000180220
ENSDART00000189937 |
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr23_+_27778670 | 1.44 |
ENSDART00000053863
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr18_+_34181655 | 1.30 |
ENSDART00000130831
ENSDART00000109535 |
gmps
|
guanine monophosphate synthase |
| chr5_+_27267186 | 1.29 |
ENSDART00000182238
ENSDART00000087857 |
unc5db
|
unc-5 netrin receptor Db |
| chr19_-_30510930 | 1.26 |
ENSDART00000088760
ENSDART00000181043 |
bag6l
|
BCL2 associated athanogene 6, like |
| chr11_+_1657221 | 1.25 |
ENSDART00000164394
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr7_+_3872189 | 1.13 |
ENSDART00000064221
|
si:dkey-88n24.5
|
si:dkey-88n24.5 |
| chr16_-_39859119 | 1.10 |
ENSDART00000138388
|
rbms3
|
RNA binding motif, single stranded interacting protein |
| chr8_+_48943009 | 0.99 |
ENSDART00000180763
|
rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr18_-_47103131 | 0.96 |
ENSDART00000188553
|
CABZ01039304.1
|
|
| chr19_-_31707892 | 0.94 |
ENSDART00000088427
|
ripor2
|
RHO family interacting cell polarization regulator 2 |
| chr8_+_48942470 | 0.92 |
ENSDART00000005464
ENSDART00000132035 |
rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr8_-_11004726 | 0.66 |
ENSDART00000192594
ENSDART00000020116 |
trim33
|
tripartite motif containing 33 |
| chr2_+_1988036 | 0.53 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr25_-_19574146 | 0.50 |
ENSDART00000156811
|
si:ch211-59o9.10
|
si:ch211-59o9.10 |
| chr5_+_25736979 | 0.48 |
ENSDART00000175959
|
abhd17b
|
abhydrolase domain containing 17B |
| chr1_+_16548733 | 0.48 |
ENSDART00000048855
|
mtus1b
|
microtubule associated tumor suppressor 1b |
| chr23_-_35347714 | 0.47 |
ENSDART00000161770
ENSDART00000165615 |
cpne9
|
copine family member IX |
| chr11_-_26180714 | 0.46 |
ENSDART00000173597
|
kaznb
|
kazrin, periplakin interacting protein b |
| chr22_+_110158 | 0.46 |
ENSDART00000143698
|
prkar2ab
|
protein kinase, cAMP-dependent, regulatory, type II, alpha, B |
| chr25_+_258883 | 0.46 |
ENSDART00000155256
|
zgc:92481
|
zgc:92481 |
| chr21_+_39941559 | 0.34 |
ENSDART00000189718
ENSDART00000160875 ENSDART00000135235 |
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr24_+_30695120 | 0.33 |
ENSDART00000178649
|
zgc:174719
|
zgc:174719 |
| chr1_+_26467071 | 0.29 |
ENSDART00000112329
ENSDART00000159318 ENSDART00000193833 ENSDART00000011809 |
uso1
|
USO1 vesicle transport factor |
| chr11_+_13067028 | 0.28 |
ENSDART00000186126
|
zfyve9b
|
zinc finger, FYVE domain containing 9b |
| chr20_-_20312789 | 0.27 |
ENSDART00000114779
|
si:ch211-212g7.6
|
si:ch211-212g7.6 |
| chr21_-_14773692 | 0.22 |
ENSDART00000142145
|
glulc
|
glutamate-ammonia ligase (glutamine synthase) c |
| chr25_+_36405021 | 0.22 |
ENSDART00000152801
|
ankrd27
|
ankyrin repeat domain 27 (VPS9 domain) |
| chr1_-_25486471 | 0.10 |
ENSDART00000134200
ENSDART00000141892 ENSDART00000102501 |
arfip1
|
ADP-ribosylation factor interacting protein 1 (arfaptin 1) |
| chr14_+_39258569 | 0.08 |
ENSDART00000103298
|
diaph2
|
diaphanous-related formin 2 |
| chr7_+_22680560 | 0.01 |
ENSDART00000133761
|
ponzr4
|
plac8 onzin related protein 4 |
| chr1_-_53476758 | 0.00 |
ENSDART00000074221
|
b3gnt2a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2a |
Network of associatons between targets according to the STRING database.
First level regulatory network of tfap2e
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 1.0 | 3.1 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 1.0 | 15.1 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.7 | 2.9 | GO:1902260 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.5 | 13.0 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.5 | 3.9 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.4 | 2.0 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.4 | 3.4 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.3 | 2.8 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.3 | 0.9 | GO:1990868 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.3 | 1.9 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.2 | 1.5 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.2 | 4.4 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.2 | 5.0 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.2 | 2.3 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.2 | 1.4 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 3.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 1.3 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.1 | 1.3 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 1.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 2.4 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 5.4 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.1 | 3.6 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.1 | 2.1 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 1.4 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 0.7 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.1 | 4.1 | GO:0050906 | detection of stimulus involved in sensory perception(GO:0050906) |
| 0.1 | 0.3 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.1 | 4.0 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 2.7 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.5 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 1.1 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.0 | 4.2 | GO:1990778 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 0.3 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 2.2 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 2.9 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.2 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 10.0 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
| 0.0 | 5.7 | GO:0006897 | endocytosis(GO:0006897) |
| 0.0 | 2.1 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.2 | 2.9 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 4.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.2 | 1.3 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.2 | 7.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 15.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 2.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 3.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 4.4 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.1 | 2.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 3.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 4.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 1.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 4.1 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 0.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.9 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 3.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 9.9 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 2.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 3.7 | GO:0005730 | nucleolus(GO:0005730) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 13.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.9 | 3.6 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.5 | 7.8 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.4 | 15.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.3 | 4.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.3 | 5.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 0.9 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.2 | 2.9 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.2 | 4.8 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 4.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 1.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 3.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.2 | 2.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 3.1 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 2.9 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 3.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 2.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 1.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 1.4 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 4.1 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 1.3 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 1.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 1.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.3 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 2.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.5 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 1.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 4.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 4.4 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 11.9 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.2 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.5 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 4.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 2.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 15.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 2.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 1.5 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |