Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
Results for vax2
Z-value: 0.51
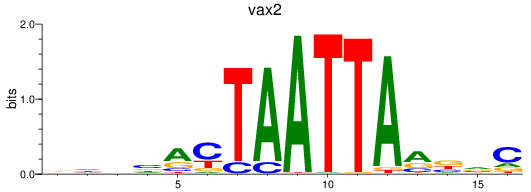
Transcription factors associated with vax2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
vax2
|
ENSDARG00000058702 | ventral anterior homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| vax2 | dr11_v1_chr7_-_8881514_8881514 | 0.03 | 7.9e-01 | Click! |
Activity profile of vax2 motif
Sorted Z-values of vax2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_29160102 | 8.68 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr6_-_46941245 | 7.77 |
ENSDART00000037875
|
igfn1.1
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 1 |
| chr5_+_37903790 | 4.43 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr12_+_3078221 | 3.76 |
ENSDART00000148835
ENSDART00000149427 |
sgca
|
sarcoglycan, alpha |
| chr10_-_34002185 | 3.11 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr6_-_46861676 | 3.05 |
ENSDART00000188712
ENSDART00000190148 |
igfn1.3
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 3 |
| chr11_-_2131280 | 2.60 |
ENSDART00000008409
|
calcoco1b
|
calcium binding and coiled-coil domain 1b |
| chr12_-_33357655 | 2.54 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr11_-_44801968 | 2.27 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr13_-_37229245 | 2.18 |
ENSDART00000140923
|
si:dkeyp-77c8.5
|
si:dkeyp-77c8.5 |
| chr14_-_858985 | 1.98 |
ENSDART00000148687
ENSDART00000149375 |
slc34a1a
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 1a |
| chr5_-_68093169 | 1.97 |
ENSDART00000051849
|
slc25a11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chr14_+_5385855 | 1.80 |
ENSDART00000031508
|
lbx2
|
ladybird homeobox 2 |
| chr14_+_45406299 | 1.76 |
ENSDART00000173142
ENSDART00000112377 |
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr11_-_6452444 | 1.72 |
ENSDART00000137879
ENSDART00000134957 ENSDART00000004483 |
larp6b
|
La ribonucleoprotein domain family, member 6b |
| chr16_-_42056137 | 1.70 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr2_+_6253246 | 1.60 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr15_-_33925851 | 1.55 |
ENSDART00000187807
ENSDART00000187780 |
mag
|
myelin associated glycoprotein |
| chr18_-_40708537 | 1.54 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr13_+_38817871 | 1.53 |
ENSDART00000187708
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr17_-_49978986 | 1.46 |
ENSDART00000154728
|
col12a1a
|
collagen, type XII, alpha 1a |
| chr7_+_42461850 | 1.45 |
ENSDART00000190350
|
adamts18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
| chr8_+_30664077 | 1.38 |
ENSDART00000138750
|
adora2aa
|
adenosine A2a receptor a |
| chr14_-_34771371 | 1.26 |
ENSDART00000160598
ENSDART00000150413 ENSDART00000168910 |
ablim3
|
actin binding LIM protein family, member 3 |
| chr21_+_31150438 | 1.26 |
ENSDART00000065366
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr10_+_42423318 | 1.25 |
ENSDART00000134282
|
npy8ar
|
neuropeptide Y receptor Y8a |
| chr6_-_7726849 | 1.23 |
ENSDART00000151511
|
slc25a38b
|
solute carrier family 25, member 38b |
| chr19_+_5480327 | 1.17 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr18_+_34225520 | 1.16 |
ENSDART00000126115
|
v2rl1
|
vomeronasal 2 receptor, l1 |
| chr13_-_35808904 | 1.15 |
ENSDART00000171667
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr1_-_55248496 | 1.11 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr15_+_34988148 | 1.09 |
ENSDART00000076269
|
ccdc105
|
coiled-coil domain containing 105 |
| chr9_-_43538328 | 1.08 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr1_+_19303241 | 1.07 |
ENSDART00000129970
|
si:dkeyp-118a3.2
|
si:dkeyp-118a3.2 |
| chr6_+_7421898 | 1.06 |
ENSDART00000043946
|
ccdc65
|
coiled-coil domain containing 65 |
| chr18_+_27821856 | 1.00 |
ENSDART00000131712
|
si:ch211-222m18.4
|
si:ch211-222m18.4 |
| chr8_-_53044300 | 0.99 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr21_+_42930558 | 0.99 |
ENSDART00000135234
|
stk32a
|
serine/threonine kinase 32A |
| chr1_-_33351780 | 0.94 |
ENSDART00000178996
|
gyg2
|
glycogenin 2 |
| chr16_+_13818500 | 0.93 |
ENSDART00000135245
|
flcn
|
folliculin |
| chr1_+_21731382 | 0.93 |
ENSDART00000054395
|
pax5
|
paired box 5 |
| chr16_-_32837806 | 0.90 |
ENSDART00000003997
|
MCHR2
|
si:dkey-165n16.5 |
| chr8_+_45186989 | 0.89 |
ENSDART00000143861
|
si:ch211-220m6.4
|
si:ch211-220m6.4 |
| chr4_-_58978527 | 0.89 |
ENSDART00000150408
|
si:ch211-64i20.5_2
|
si:ch211-64i20.5_2 |
| chr25_+_10913725 | 0.89 |
ENSDART00000153472
|
mhc1lja
|
major histocompatibility complex class I LJA |
| chr21_+_31150773 | 0.88 |
ENSDART00000126205
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr23_+_4709607 | 0.88 |
ENSDART00000166503
ENSDART00000158752 ENSDART00000163860 ENSDART00000172739 |
raf1a
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a Raf-1 proto-oncogene, serine/threonine kinase a |
| chr22_-_209741 | 0.88 |
ENSDART00000171954
|
ora1
|
olfactory receptor class A related 1 |
| chr22_+_7738966 | 0.88 |
ENSDART00000147073
|
si:ch73-44m9.5
|
si:ch73-44m9.5 |
| chr16_-_15387459 | 0.87 |
ENSDART00000053754
|
has2
|
hyaluronan synthase 2 |
| chr14_-_4556896 | 0.85 |
ENSDART00000044678
ENSDART00000192863 |
GABRA2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr7_+_54642005 | 0.85 |
ENSDART00000171864
|
fgf19
|
fibroblast growth factor 19 |
| chr21_-_31290582 | 0.83 |
ENSDART00000065362
|
ca4c
|
carbonic anhydrase IV c |
| chr15_-_35134918 | 0.82 |
ENSDART00000157196
|
si:ch211-272b8.7
|
si:ch211-272b8.7 |
| chr14_-_8724290 | 0.80 |
ENSDART00000161171
|
pimr56
|
Pim proto-oncogene, serine/threonine kinase, related 56 |
| chr8_+_28695914 | 0.80 |
ENSDART00000033386
|
ocstamp
|
osteoclast stimulatory transmembrane protein |
| chr4_+_44954486 | 0.79 |
ENSDART00000150875
|
si:dkeyp-100h4.8
|
si:dkeyp-100h4.8 |
| chr16_-_11798994 | 0.78 |
ENSDART00000135408
|
cnfn
|
cornifelin |
| chr25_-_13490744 | 0.78 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr17_+_8799661 | 0.77 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr15_-_4528326 | 0.77 |
ENSDART00000158122
ENSDART00000155619 ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr1_+_49668423 | 0.76 |
ENSDART00000150880
|
tsga10
|
testis specific, 10 |
| chr2_+_53204750 | 0.75 |
ENSDART00000163644
|
zgc:165603
|
zgc:165603 |
| chr23_+_30827959 | 0.74 |
ENSDART00000033429
|
npbwr2a
|
neuropeptides B/W receptor 2a |
| chr13_-_38039871 | 0.73 |
ENSDART00000140645
|
CR456624.1
|
|
| chr13_+_255067 | 0.71 |
ENSDART00000102505
|
foxg1d
|
forkhead box G1d |
| chr7_-_12464412 | 0.70 |
ENSDART00000178723
|
adamtsl3
|
ADAMTS-like 3 |
| chr23_+_238893 | 0.70 |
ENSDART00000138198
|
PDZD4
|
si:ch73-162j3.4 |
| chr21_+_32820175 | 0.68 |
ENSDART00000076903
|
adra2db
|
adrenergic, alpha-2D-, receptor b |
| chr11_-_2838699 | 0.68 |
ENSDART00000066189
|
lhfpl5a
|
LHFPL tetraspan subfamily member 5a |
| chr20_+_28861629 | 0.68 |
ENSDART00000187274
ENSDART00000047826 |
slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr6_-_59942335 | 0.66 |
ENSDART00000168416
|
fbxl3b
|
F-box and leucine-rich repeat protein 3b |
| chr11_+_705727 | 0.63 |
ENSDART00000165366
|
timp4.2
|
TIMP metallopeptidase inhibitor 4, tandem duplicate 2 |
| chr4_-_13548806 | 0.62 |
ENSDART00000067155
|
il22
|
interleukin 22 |
| chr6_+_27151940 | 0.61 |
ENSDART00000088364
|
kif1aa
|
kinesin family member 1Aa |
| chr13_-_36798204 | 0.61 |
ENSDART00000012357
|
sav1
|
salvador family WW domain containing protein 1 |
| chr20_-_43663494 | 0.61 |
ENSDART00000144564
|
BX470188.1
|
|
| chr2_+_10007113 | 0.61 |
ENSDART00000155213
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr14_-_30587814 | 0.61 |
ENSDART00000144912
ENSDART00000149714 |
tmem265
|
transmembrane protein 265 |
| chr1_-_19502322 | 0.60 |
ENSDART00000181888
ENSDART00000044030 |
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr24_+_26134209 | 0.60 |
ENSDART00000038824
|
tmtopsb
|
teleost multiple tissue opsin b |
| chr20_-_28800999 | 0.59 |
ENSDART00000049462
|
rab15
|
RAB15, member RAS oncogene family |
| chr2_-_9818640 | 0.59 |
ENSDART00000139499
ENSDART00000165548 ENSDART00000012442 ENSDART00000046587 |
ap2m1b
|
adaptor-related protein complex 2, mu 1 subunit, b |
| chr4_-_2975461 | 0.57 |
ENSDART00000150794
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr10_+_38560032 | 0.57 |
ENSDART00000177197
|
LO018336.1
|
|
| chr15_+_45640906 | 0.54 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr12_+_47698356 | 0.54 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr12_+_48803098 | 0.52 |
ENSDART00000074768
|
ppifb
|
peptidylprolyl isomerase Fb |
| chr3_-_34034498 | 0.52 |
ENSDART00000151145
|
ighv11-2
|
immunoglobulin heavy variable 11-2 |
| chr11_-_2478374 | 0.51 |
ENSDART00000173205
|
si:ch73-267c23.10
|
si:ch73-267c23.10 |
| chr22_+_18477934 | 0.50 |
ENSDART00000132684
|
cilp2
|
cartilage intermediate layer protein 2 |
| chr15_+_31303355 | 0.50 |
ENSDART00000143423
|
or116-2
|
odorant receptor, family F, subfamily 116, member 2 |
| chr7_-_60156409 | 0.50 |
ENSDART00000006802
|
cct7
|
chaperonin containing TCP1, subunit 7 (eta) |
| chr10_+_16584382 | 0.48 |
ENSDART00000112039
|
CR790388.1
|
|
| chr2_+_22808946 | 0.48 |
ENSDART00000133250
|
aire
|
autoimmune regulator |
| chr17_+_3379673 | 0.46 |
ENSDART00000176354
|
sntg2
|
syntrophin, gamma 2 |
| chr17_-_10122204 | 0.46 |
ENSDART00000160751
|
BX088587.1
|
|
| chr13_+_40501455 | 0.46 |
ENSDART00000114985
|
hpse2
|
heparanase 2 |
| chr16_+_31804590 | 0.45 |
ENSDART00000167321
|
wnt4b
|
wingless-type MMTV integration site family, member 4b |
| chr8_+_49778486 | 0.45 |
ENSDART00000131732
|
ntrk2a
|
neurotrophic tyrosine kinase, receptor, type 2a |
| chr24_+_37640626 | 0.43 |
ENSDART00000008047
|
wdr24
|
WD repeat domain 24 |
| chr8_+_16025554 | 0.43 |
ENSDART00000110171
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr14_+_29941445 | 0.41 |
ENSDART00000181761
|
fam149a
|
family with sequence similarity 149 member A |
| chr4_+_966061 | 0.41 |
ENSDART00000122535
|
rpap3
|
RNA polymerase II associated protein 3 |
| chr13_-_29420885 | 0.39 |
ENSDART00000024225
|
chata
|
choline O-acetyltransferase a |
| chr20_-_7133546 | 0.39 |
ENSDART00000160910
|
si:ch211-121a2.4
|
si:ch211-121a2.4 |
| chr1_-_15797663 | 0.38 |
ENSDART00000177122
|
SGCZ
|
sarcoglycan zeta |
| chr18_+_28106139 | 0.38 |
ENSDART00000089615
|
kiaa1549lb
|
KIAA1549-like b |
| chr4_+_9400012 | 0.38 |
ENSDART00000191960
|
tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr1_-_17715493 | 0.37 |
ENSDART00000133027
|
si:dkey-256e7.8
|
si:dkey-256e7.8 |
| chr19_+_3213914 | 0.35 |
ENSDART00000193144
|
zgc:86598
|
zgc:86598 |
| chr11_+_31864921 | 0.34 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr11_-_40728380 | 0.34 |
ENSDART00000023745
|
ccdc114
|
coiled-coil domain containing 114 |
| chr4_+_77943184 | 0.34 |
ENSDART00000159094
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr6_+_24398907 | 0.33 |
ENSDART00000167482
|
tgfbr3
|
transforming growth factor, beta receptor III |
| chr21_-_13972745 | 0.33 |
ENSDART00000143874
|
akna
|
AT-hook transcription factor |
| chrM_+_12897 | 0.32 |
ENSDART00000093622
|
mt-nd5
|
NADH dehydrogenase 5, mitochondrial |
| chr20_-_9278252 | 0.32 |
ENSDART00000136079
|
syt14b
|
synaptotagmin XIVb |
| chr17_-_200316 | 0.32 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr20_+_28861435 | 0.30 |
ENSDART00000142678
|
slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr5_-_51619742 | 0.29 |
ENSDART00000188537
|
otpb
|
orthopedia homeobox b |
| chr3_+_17537352 | 0.26 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr10_+_11261576 | 0.26 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr15_-_22074315 | 0.26 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr23_-_15216654 | 0.25 |
ENSDART00000131649
|
sulf2b
|
sulfatase 2b |
| chr10_-_2788668 | 0.24 |
ENSDART00000131749
ENSDART00000124356 ENSDART00000085031 |
ash2l
|
ash2 (absent, small, or homeotic)-like (Drosophila) |
| chr8_+_23738122 | 0.24 |
ENSDART00000062983
|
rpl10a
|
ribosomal protein L10a |
| chr9_-_34937025 | 0.24 |
ENSDART00000137888
|
cdc16
|
cell division cycle 16 homolog (S. cerevisiae) |
| chr11_+_33312601 | 0.24 |
ENSDART00000188024
|
cntnap5l
|
contactin associated protein-like 5 like |
| chr2_+_105748 | 0.23 |
ENSDART00000169601
|
CABZ01098670.1
|
|
| chr12_+_20587179 | 0.23 |
ENSDART00000170127
|
arsg
|
arylsulfatase G |
| chr3_-_21280373 | 0.22 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr2_+_50608099 | 0.19 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr14_+_40874608 | 0.19 |
ENSDART00000168448
|
si:ch211-106m9.1
|
si:ch211-106m9.1 |
| chr22_+_23359369 | 0.18 |
ENSDART00000170886
|
dennd1b
|
DENN/MADD domain containing 1B |
| chr24_-_37640705 | 0.17 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr4_-_48957129 | 0.17 |
ENSDART00000150426
|
znf1064
|
zinc finger protein 1064 |
| chr11_-_42554290 | 0.16 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr7_-_30174882 | 0.16 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr2_+_53720028 | 0.16 |
ENSDART00000170799
|
ctnnbl1
|
catenin, beta like 1 |
| chr24_+_12894282 | 0.12 |
ENSDART00000061301
|
si:dkeyp-28d2.4
|
si:dkeyp-28d2.4 |
| chr5_-_16472719 | 0.12 |
ENSDART00000162071
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr16_-_20932896 | 0.12 |
ENSDART00000180646
|
tax1bp1b
|
Tax1 (human T-cell leukemia virus type I) binding protein 1b |
| chr17_+_30369107 | 0.10 |
ENSDART00000182286
ENSDART00000139091 |
greb1
|
growth regulation by estrogen in breast cancer 1 |
| chr23_-_2901167 | 0.09 |
ENSDART00000165955
ENSDART00000190616 |
zhx3
|
zinc fingers and homeoboxes 3 |
| chr23_-_35790235 | 0.09 |
ENSDART00000142369
ENSDART00000141141 ENSDART00000011004 |
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr20_-_9095105 | 0.08 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr16_+_13818743 | 0.07 |
ENSDART00000090191
|
flcn
|
folliculin |
| chr8_-_53680653 | 0.05 |
ENSDART00000164739
|
wnt5a
|
wingless-type MMTV integration site family, member 5a |
| chr23_+_2906031 | 0.05 |
ENSDART00000109304
|
c23h20orf24
|
c23h20orf24 homolog (H. sapiens) |
| chr8_+_18011522 | 0.05 |
ENSDART00000136756
|
ssbp3b
|
single stranded DNA binding protein 3b |
| chr8_+_2543525 | 0.04 |
ENSDART00000129569
ENSDART00000128227 |
gle1
|
GLE1 RNA export mediator homolog (yeast) |
| chr9_-_44295071 | 0.03 |
ENSDART00000011837
|
neurod1
|
neuronal differentiation 1 |
| chr8_+_23521974 | 0.03 |
ENSDART00000188130
ENSDART00000129378 |
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr4_-_30422325 | 0.02 |
ENSDART00000158444
|
znf1114
|
zinc finger protein 1114 |
| chr7_+_9873750 | 0.01 |
ENSDART00000173348
ENSDART00000172813 |
lins1
|
lines homolog 1 |
| chr10_-_11261386 | 0.01 |
ENSDART00000189946
|
ptbp3
|
polypyrimidine tract binding protein 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of vax2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.4 | 2.0 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.4 | 1.8 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.3 | 1.4 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.3 | 2.0 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.3 | 1.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.3 | 0.9 | GO:0003091 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.3 | 0.8 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.2 | 1.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.2 | 4.0 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 3.3 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 0.5 | GO:0032602 | chemokine production(GO:0032602) negative T cell selection(GO:0043383) thymocyte migration(GO:0072679) |
| 0.2 | 0.6 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 0.4 | GO:1990416 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 1.0 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 1.1 | GO:0003352 | regulation of cilium movement(GO:0003352) |
| 0.1 | 0.7 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.1 | 8.7 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.1 | 0.4 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.1 | 0.6 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.9 | GO:0060005 | reflex(GO:0060004) vestibular reflex(GO:0060005) vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.1 | 2.2 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 0.3 | GO:0015990 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.1 | 0.9 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.1 | 1.5 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.1 | 1.0 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.5 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.3 | GO:0042755 | eating behavior(GO:0042755) |
| 0.1 | 0.9 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 1.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.3 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.0 | 0.5 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.5 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.6 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 2.6 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 2.5 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.8 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.5 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.3 | GO:0001765 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.7 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.6 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 1.1 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.3 | GO:0051967 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.9 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.7 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.0 | 0.2 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.9 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.6 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 2.0 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 1.6 | GO:1902284 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.0 | 0.4 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 1.0 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.5 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 2.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.8 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 4.0 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 2.0 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 1.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 1.1 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 0.5 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 1.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.2 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.5 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.3 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 1.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.3 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 8.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.4 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.5 | GO:0030496 | midbody(GO:0030496) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 8.7 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.4 | 2.0 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.3 | 2.0 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.3 | 0.9 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.3 | 0.9 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.3 | 2.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.2 | 2.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 1.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.2 | 3.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 1.0 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.4 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.1 | 1.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.7 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 0.8 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.1 | 1.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 1.6 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.7 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.6 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 1.3 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 4.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.6 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 2.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 4.0 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.4 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.1 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 2.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.6 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 1.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.5 | GO:0045182 | translation regulator activity(GO:0045182) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.8 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.9 | PID FGF PATHWAY | FGF signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 4.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 1.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 0.9 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 2.0 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 0.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 0.9 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 0.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.2 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |