Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
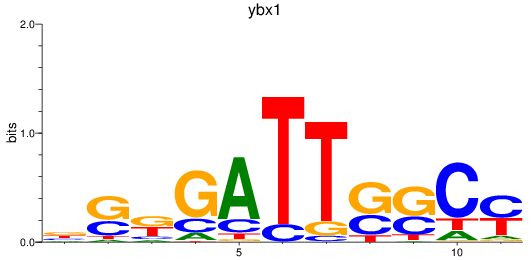
Results for ybx1
Z-value: 1.09
Transcription factors associated with ybx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ybx1
|
ENSDARG00000004757 | Y box binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ybx1 | dr11_v1_chr8_-_47152001_47152086 | -0.59 | 2.6e-10 | Click! |
Activity profile of ybx1 motif
Sorted Z-values of ybx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_44883805 | 30.48 |
ENSDART00000182805
|
si:ch73-361h17.1
|
si:ch73-361h17.1 |
| chr22_+_38173960 | 19.76 |
ENSDART00000010537
|
cp
|
ceruloplasmin |
| chr25_-_4148719 | 16.47 |
ENSDART00000112880
ENSDART00000023278 |
fads2
|
fatty acid desaturase 2 |
| chr18_+_62932 | 12.24 |
ENSDART00000052638
|
slc27a2a
|
solute carrier family 27 (fatty acid transporter), member 2a |
| chr13_+_2394534 | 10.20 |
ENSDART00000172535
ENSDART00000006990 |
elovl5
|
ELOVL fatty acid elongase 5 |
| chr1_-_20928772 | 9.06 |
ENSDART00000078277
|
msmo1
|
methylsterol monooxygenase 1 |
| chr10_+_6010570 | 7.96 |
ENSDART00000190025
ENSDART00000163680 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr11_-_44543082 | 7.86 |
ENSDART00000099568
|
gpr137bb
|
G protein-coupled receptor 137Bb |
| chr10_+_10788811 | 7.78 |
ENSDART00000101077
ENSDART00000139143 |
ptgdsa
|
prostaglandin D2 synthase a |
| chr9_+_27411502 | 7.65 |
ENSDART00000143994
|
si:dkey-193n17.9
|
si:dkey-193n17.9 |
| chr18_-_3166726 | 7.54 |
ENSDART00000165002
|
aqp11
|
aquaporin 11 |
| chr13_+_2394264 | 7.44 |
ENSDART00000168595
|
elovl5
|
ELOVL fatty acid elongase 5 |
| chr6_+_3710865 | 7.03 |
ENSDART00000170781
|
phospho2
|
phosphatase, orphan 2 |
| chr6_-_34958274 | 6.74 |
ENSDART00000113097
|
hsd17b7
|
hydroxysteroid (17-beta) dehydrogenase 7 |
| chr6_-_35446110 | 6.47 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr25_-_19433244 | 6.28 |
ENSDART00000154778
|
map1ab
|
microtubule-associated protein 1Ab |
| chr16_-_39477746 | 6.23 |
ENSDART00000102525
|
stt3b
|
STT3B, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr19_-_30562648 | 6.14 |
ENSDART00000171006
|
hpcal4
|
hippocalcin like 4 |
| chr22_+_17205608 | 6.12 |
ENSDART00000181267
|
rab3b
|
RAB3B, member RAS oncogene family |
| chr4_+_11375894 | 6.08 |
ENSDART00000190471
ENSDART00000143963 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr16_-_39477509 | 5.81 |
ENSDART00000191330
|
stt3b
|
STT3B, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr24_+_17334682 | 5.75 |
ENSDART00000018868
|
pdia4
|
protein disulfide isomerase family A, member 4 |
| chr1_-_18803919 | 5.46 |
ENSDART00000020970
|
pgm2
|
phosphoglucomutase 2 |
| chr24_+_7708652 | 5.39 |
ENSDART00000066816
|
btr33
|
bloodthirsty-related gene family, member 33 |
| chr15_+_43166511 | 5.38 |
ENSDART00000011737
|
flj13639
|
flj13639 |
| chr16_-_44349845 | 5.36 |
ENSDART00000170932
|
rims2a
|
regulating synaptic membrane exocytosis 2a |
| chr17_+_23556764 | 5.25 |
ENSDART00000146787
|
pank1a
|
pantothenate kinase 1a |
| chr20_-_52939501 | 5.24 |
ENSDART00000166508
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr20_-_7176809 | 5.19 |
ENSDART00000012247
ENSDART00000125019 |
dhcr24
|
24-dehydrocholesterol reductase |
| chr6_-_52156427 | 4.92 |
ENSDART00000082821
|
rims4
|
regulating synaptic membrane exocytosis 4 |
| chr8_-_39884359 | 4.80 |
ENSDART00000131372
|
mlec
|
malectin |
| chr3_+_17951790 | 4.73 |
ENSDART00000164663
|
aclya
|
ATP citrate lyase a |
| chr3_+_59051503 | 4.67 |
ENSDART00000160767
|
rasd4
|
rasd family member 4 |
| chr2_+_24868010 | 4.66 |
ENSDART00000078838
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr22_-_36875264 | 4.61 |
ENSDART00000137548
|
kng1
|
kininogen 1 |
| chr10_+_7703251 | 4.58 |
ENSDART00000165134
|
ggcx
|
gamma-glutamyl carboxylase |
| chr15_-_19250543 | 4.42 |
ENSDART00000092705
ENSDART00000138895 |
igsf9ba
|
immunoglobulin superfamily, member 9Ba |
| chr18_+_27515640 | 4.31 |
ENSDART00000181593
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr4_-_28158335 | 4.12 |
ENSDART00000134605
|
gramd4a
|
GRAM domain containing 4a |
| chr23_+_45579497 | 4.07 |
ENSDART00000110381
|
egr4
|
early growth response 4 |
| chr21_+_19070921 | 4.00 |
ENSDART00000029874
|
nkx6.1
|
NK6 homeobox 1 |
| chr24_+_34606966 | 3.87 |
ENSDART00000105477
|
lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr15_-_20731297 | 3.84 |
ENSDART00000114464
|
tpst1
|
tyrosylprotein sulfotransferase 1 |
| chr14_-_16476863 | 3.83 |
ENSDART00000089021
|
canx
|
calnexin |
| chr16_+_46695777 | 3.77 |
ENSDART00000169767
|
rab25b
|
RAB25, member RAS oncogene family b |
| chr7_+_1521834 | 3.74 |
ENSDART00000174007
|
si:cabz01102082.1
|
si:cabz01102082.1 |
| chr21_+_244503 | 3.73 |
ENSDART00000162889
|
stard4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr23_+_19564392 | 3.71 |
ENSDART00000144746
|
atp6ap1lb
|
ATPase H+ transporting accessory protein 1 like b |
| chr4_-_5247335 | 3.71 |
ENSDART00000050221
|
atp6v1e1b
|
ATPase H+ transporting V1 subunit E1b |
| chr3_-_13147310 | 3.59 |
ENSDART00000160840
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr11_-_15297129 | 3.55 |
ENSDART00000167985
ENSDART00000163031 ENSDART00000165010 ENSDART00000171250 ENSDART00000010684 ENSDART00000191714 ENSDART00000191164 |
rpn2
|
ribophorin II |
| chr3_-_32541033 | 3.39 |
ENSDART00000151476
ENSDART00000055324 |
rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr9_-_1604601 | 3.31 |
ENSDART00000143130
|
agps
|
alkylglycerone phosphate synthase |
| chr15_-_3094516 | 3.26 |
ENSDART00000179719
|
slitrk5a
|
SLIT and NTRK-like family, member 5a |
| chr20_+_16743056 | 3.26 |
ENSDART00000050308
|
calm1b
|
calmodulin 1b |
| chr3_+_3810919 | 3.16 |
ENSDART00000056035
|
FQ311927.1
|
|
| chr17_+_31221761 | 3.14 |
ENSDART00000155580
|
ccdc32
|
coiled-coil domain containing 32 |
| chr18_+_7073130 | 3.13 |
ENSDART00000101216
ENSDART00000148947 |
si:dkey-88e18.2
|
si:dkey-88e18.2 |
| chr2_+_24867534 | 3.12 |
ENSDART00000158050
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr24_-_39772045 | 3.11 |
ENSDART00000087441
|
GFOD1
|
si:ch211-276f18.2 |
| chr21_+_3897680 | 3.10 |
ENSDART00000170653
|
dolpp1
|
dolichyldiphosphatase 1 |
| chr25_+_25508495 | 3.08 |
ENSDART00000150719
|
phrf1
|
PHD and ring finger domains 1 |
| chr24_+_24923166 | 3.08 |
ENSDART00000065288
|
pcyt1ba
|
phosphate cytidylyltransferase 1, choline, beta a |
| chr12_+_1469327 | 3.03 |
ENSDART00000059143
|
usp22
|
ubiquitin specific peptidase 22 |
| chr21_+_5888641 | 3.03 |
ENSDART00000091331
|
prodha
|
proline dehydrogenase (oxidase) 1a |
| chr1_-_50527964 | 2.99 |
ENSDART00000024984
|
papss1
|
3'-phosphoadenosine 5'-phosphosulfate synthase 1 |
| chr2_+_37480669 | 2.83 |
ENSDART00000029801
|
sppl2
|
signal peptide peptidase-like 2 |
| chr1_+_54737353 | 2.82 |
ENSDART00000130675
ENSDART00000162075 |
pi4k2a
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr21_+_944715 | 2.76 |
ENSDART00000144766
|
nars
|
asparaginyl-tRNA synthetase |
| chr1_+_24076243 | 2.76 |
ENSDART00000014608
|
mab21l2
|
mab-21-like 2 |
| chr11_-_30352333 | 2.71 |
ENSDART00000030794
|
tmem169a
|
transmembrane protein 169a |
| chr9_+_2452672 | 2.65 |
ENSDART00000193993
|
chn1
|
chimerin 1 |
| chr8_+_54081819 | 2.56 |
ENSDART00000005857
ENSDART00000161795 |
prickle2a
|
prickle homolog 2a |
| chr21_-_26406244 | 2.56 |
ENSDART00000137312
ENSDART00000077200 |
eif4ebp3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr12_+_46960579 | 2.54 |
ENSDART00000149032
|
oat
|
ornithine aminotransferase |
| chr20_-_3403033 | 2.41 |
ENSDART00000092264
|
rev3l
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr21_+_34981263 | 2.41 |
ENSDART00000132711
|
rbm11
|
RNA binding motif protein 11 |
| chr5_+_20030414 | 2.39 |
ENSDART00000181430
ENSDART00000047841 ENSDART00000182813 |
sgsm1a
|
small G protein signaling modulator 1a |
| chr6_-_25952848 | 2.37 |
ENSDART00000076997
ENSDART00000148748 |
lmo4b
|
LIM domain only 4b |
| chr14_-_12020653 | 2.32 |
ENSDART00000106654
|
znf711
|
zinc finger protein 711 |
| chr23_+_6795531 | 2.31 |
ENSDART00000092131
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr23_+_2778813 | 2.30 |
ENSDART00000142621
|
top1
|
DNA topoisomerase I |
| chr21_-_12119711 | 2.29 |
ENSDART00000131538
|
celf4
|
CUGBP, Elav-like family member 4 |
| chr3_-_58543658 | 2.29 |
ENSDART00000042386
|
unm_sa1261
|
un-named sa1261 |
| chr1_+_43686251 | 2.29 |
ENSDART00000074604
ENSDART00000137791 |
cisd2
|
CDGSH iron sulfur domain 2 |
| chr4_+_4079418 | 2.23 |
ENSDART00000028016
|
waslb
|
Wiskott-Aldrich syndrome-like b |
| chr11_-_42554290 | 2.23 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr5_-_16983336 | 2.22 |
ENSDART00000038740
|
galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr15_-_19677511 | 2.16 |
ENSDART00000043743
|
si:dkey-4p15.3
|
si:dkey-4p15.3 |
| chr16_-_32672883 | 2.15 |
ENSDART00000124515
ENSDART00000190920 ENSDART00000188776 |
pnisr
|
PNN-interacting serine/arginine-rich protein |
| chr2_-_4797512 | 2.13 |
ENSDART00000160765
|
tnk2b
|
tyrosine kinase, non-receptor, 2b |
| chr19_+_42806812 | 2.13 |
ENSDART00000108775
ENSDART00000151653 |
ubp1
|
upstream binding protein 1 |
| chr16_-_12095144 | 2.12 |
ENSDART00000145106
|
pex5
|
peroxisomal biogenesis factor 5 |
| chr1_+_51386649 | 2.09 |
ENSDART00000152289
|
atg4da
|
autophagy related 4D, cysteine peptidase a |
| chr11_-_18604317 | 2.09 |
ENSDART00000182081
|
zmynd8
|
zinc finger, MYND-type containing 8 |
| chr11_-_15296805 | 2.06 |
ENSDART00000124968
|
rpn2
|
ribophorin II |
| chr11_-_18604542 | 2.05 |
ENSDART00000171183
|
zmynd8
|
zinc finger, MYND-type containing 8 |
| chr15_+_41836104 | 1.99 |
ENSDART00000137434
|
pxylp1
|
2-phosphoxylose phosphatase 1 |
| chr23_-_10786400 | 1.96 |
ENSDART00000055038
|
rybpa
|
RING1 and YY1 binding protein a |
| chr5_+_26138313 | 1.96 |
ENSDART00000010041
|
dhfr
|
dihydrofolate reductase |
| chr22_-_12693833 | 1.92 |
ENSDART00000129768
ENSDART00000044574 |
adarb1a
|
adenosine deaminase, RNA-specific, B1a |
| chr8_-_1266181 | 1.88 |
ENSDART00000148654
ENSDART00000149924 |
cdc14b
|
cell division cycle 14B |
| chr3_-_21062706 | 1.88 |
ENSDART00000155605
ENSDART00000153686 ENSDART00000157168 ENSDART00000156614 ENSDART00000155743 ENSDART00000156275 |
fam57ba
|
family with sequence similarity 57, member Ba |
| chr20_-_53366137 | 1.86 |
ENSDART00000146001
|
wasf1
|
WAS protein family, member 1 |
| chr5_-_2721686 | 1.85 |
ENSDART00000169404
|
hspa5
|
heat shock protein 5 |
| chr12_+_1469090 | 1.84 |
ENSDART00000183637
|
usp22
|
ubiquitin specific peptidase 22 |
| chr18_+_40364732 | 1.80 |
ENSDART00000123661
ENSDART00000130847 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr18_+_22606259 | 1.78 |
ENSDART00000128965
|
bcar1
|
breast cancer anti-estrogen resistance 1 |
| chr1_+_25801648 | 1.77 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr22_-_16180467 | 1.75 |
ENSDART00000171331
ENSDART00000185607 |
vcam1b
|
vascular cell adhesion molecule 1b |
| chr4_-_9196291 | 1.72 |
ENSDART00000153963
|
hcfc2
|
host cell factor C2 |
| chr11_+_14321113 | 1.71 |
ENSDART00000039822
ENSDART00000137347 ENSDART00000132997 |
ptbp1b
|
polypyrimidine tract binding protein 1b |
| chr3_-_37585863 | 1.70 |
ENSDART00000149762
|
arf2a
|
ADP-ribosylation factor 2a |
| chr12_+_38807604 | 1.68 |
ENSDART00000155563
|
abca5
|
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr9_+_22677503 | 1.67 |
ENSDART00000131429
ENSDART00000080005 ENSDART00000101756 ENSDART00000138148 |
itgb5
|
integrin, beta 5 |
| chr19_+_14109348 | 1.67 |
ENSDART00000159015
|
zgc:175136
|
zgc:175136 |
| chr13_+_8892784 | 1.65 |
ENSDART00000075054
ENSDART00000143705 |
thada
|
thyroid adenoma associated |
| chr18_-_2549198 | 1.64 |
ENSDART00000186516
|
CABZ01070631.1
|
|
| chr6_+_53349966 | 1.64 |
ENSDART00000167079
|
si:ch211-161c3.5
|
si:ch211-161c3.5 |
| chr1_-_20271138 | 1.62 |
ENSDART00000185931
|
ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr4_+_29967255 | 1.59 |
ENSDART00000150383
|
znf1128
|
zinc finger protein 1128 |
| chr13_-_30700460 | 1.56 |
ENSDART00000139073
|
rassf4
|
Ras association (RalGDS/AF-6) domain family member 4 |
| chr2_+_29249204 | 1.55 |
ENSDART00000168957
|
cdh18a
|
cadherin 18, type 2a |
| chr17_-_43287290 | 1.54 |
ENSDART00000156885
|
EML5
|
si:dkey-1f12.3 |
| chr19_+_770300 | 1.49 |
ENSDART00000062518
|
gstr
|
glutathione S-transferase rho |
| chr1_-_9644630 | 1.48 |
ENSDART00000123725
ENSDART00000161164 |
ugt5b3
|
UDP glucuronosyltransferase 5 family, polypeptide B3 |
| chr12_+_40905427 | 1.39 |
ENSDART00000170526
ENSDART00000185771 ENSDART00000193945 |
CDH18
|
cadherin 18 |
| chr19_-_6134802 | 1.39 |
ENSDART00000140051
|
cica
|
capicua transcriptional repressor a |
| chr7_-_29115772 | 1.37 |
ENSDART00000076386
|
fbxl8
|
F-box and leucine-rich repeat protein 8 |
| chr3_-_61218272 | 1.36 |
ENSDART00000133091
ENSDART00000055068 |
pvalb5
|
parvalbumin 5 |
| chr19_-_26769867 | 1.36 |
ENSDART00000043776
ENSDART00000159489 ENSDART00000138675 |
prrc2a
|
proline-rich coiled-coil 2A |
| chr3_+_62353650 | 1.35 |
ENSDART00000112428
|
iqck
|
IQ motif containing K |
| chr25_-_6261693 | 1.34 |
ENSDART00000135808
|
ireb2
|
iron-responsive element binding protein 2 |
| chr6_-_51541488 | 1.32 |
ENSDART00000156336
|
si:dkey-6e2.2
|
si:dkey-6e2.2 |
| chr18_+_17600570 | 1.32 |
ENSDART00000175258
ENSDART00000151850 ENSDART00000151934 |
herpud1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr10_-_2713228 | 1.28 |
ENSDART00000123754
ENSDART00000126236 |
mier3a
|
mesoderm induction early response 1, family member 3 a |
| chr18_+_44795711 | 1.27 |
ENSDART00000110229
ENSDART00000188262 ENSDART00000139526 |
fam118b
|
family with sequence similarity 118, member B |
| chr8_+_53269657 | 1.27 |
ENSDART00000184212
|
gnb1a
|
guanine nucleotide binding protein (G protein), beta polypeptide 1a |
| chr23_+_36616717 | 1.26 |
ENSDART00000042701
ENSDART00000192980 |
pip4k2ca
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma a |
| chr12_-_35885349 | 1.25 |
ENSDART00000085662
|
cep131
|
centrosomal protein 131 |
| chr9_-_12594759 | 1.25 |
ENSDART00000021266
|
tra2b
|
transformer 2 beta homolog |
| chr7_-_56835543 | 1.24 |
ENSDART00000010322
|
senp8
|
SUMO/sentrin peptidase family member, NEDD8 specific |
| chr3_+_16664212 | 1.23 |
ENSDART00000013816
|
zgc:55558
|
zgc:55558 |
| chr20_-_40487208 | 1.21 |
ENSDART00000075070
ENSDART00000142029 |
hsf2
|
heat shock transcription factor 2 |
| chr5_+_26686279 | 1.20 |
ENSDART00000193543
|
tango2
|
transport and golgi organization 2 homolog (Drosophila) |
| chr12_-_48671612 | 1.20 |
ENSDART00000007202
|
zgc:92749
|
zgc:92749 |
| chr19_-_26770083 | 1.18 |
ENSDART00000193811
ENSDART00000174455 |
prrc2a
|
proline-rich coiled-coil 2A |
| chr1_+_6817292 | 1.18 |
ENSDART00000145822
ENSDART00000092118 |
erbb4a
|
erb-b2 receptor tyrosine kinase 4a |
| chr20_-_49889111 | 1.15 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr17_-_43286903 | 1.14 |
ENSDART00000176637
|
EML5
|
si:dkey-1f12.3 |
| chr1_+_30723677 | 1.13 |
ENSDART00000177900
|
bora
|
bora, aurora kinase A activator |
| chr23_-_41651759 | 1.13 |
ENSDART00000146808
|
si:ch73-184c24.1
|
si:ch73-184c24.1 |
| chr11_+_18612166 | 1.13 |
ENSDART00000162694
|
ncoa3
|
nuclear receptor coactivator 3 |
| chr10_+_36026576 | 1.12 |
ENSDART00000193786
|
hmgb1a
|
high mobility group box 1a |
| chr2_+_9946121 | 1.12 |
ENSDART00000100696
|
tsen15
|
TSEN15 tRNA splicing endonuclease subunit |
| chr12_-_33582382 | 1.09 |
ENSDART00000009794
ENSDART00000136617 |
tdrkh
|
tudor and KH domain containing |
| chr25_-_17918810 | 1.09 |
ENSDART00000023959
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr20_+_18703108 | 1.09 |
ENSDART00000181188
|
eif5
|
eukaryotic translation initiation factor 5 |
| chr7_+_39360797 | 1.09 |
ENSDART00000173481
|
acp2
|
acid phosphatase 2, lysosomal |
| chr3_+_62317990 | 1.08 |
ENSDART00000058719
|
znf1124
|
zinc finger protein 1124 |
| chr12_-_10674606 | 1.08 |
ENSDART00000157919
|
med24
|
mediator complex subunit 24 |
| chr16_+_4838808 | 1.07 |
ENSDART00000179363
|
ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr3_+_35498119 | 1.04 |
ENSDART00000178963
|
tnrc6a
|
trinucleotide repeat containing 6a |
| chr11_+_26375979 | 1.03 |
ENSDART00000087652
ENSDART00000171748 ENSDART00000103513 ENSDART00000165931 ENSDART00000170043 |
cpne1
rbm12
|
copine I RNA binding motif protein 12 |
| chr9_+_45227028 | 1.02 |
ENSDART00000185579
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr25_+_4541211 | 1.02 |
ENSDART00000129978
|
pnpla2
|
patatin-like phospholipase domain containing 2 |
| chr19_-_31686252 | 0.99 |
ENSDART00000131721
|
ripor2
|
RHO family interacting cell polarization regulator 2 |
| chr16_-_52646789 | 0.99 |
ENSDART00000035761
|
ubr5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr8_-_44015210 | 0.97 |
ENSDART00000186879
ENSDART00000188965 ENSDART00000001313 ENSDART00000188902 ENSDART00000185935 ENSDART00000147869 |
rimbp2
rimbp2
|
RIMS binding protein 2 RIMS binding protein 2 |
| chr23_+_6795709 | 0.96 |
ENSDART00000149136
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr13_-_856701 | 0.96 |
ENSDART00000140423
|
TMEM14A
|
zgc:163080 |
| chr21_-_5007109 | 0.96 |
ENSDART00000187042
ENSDART00000097796 ENSDART00000146766 |
rnf165a
|
ring finger protein 165a |
| chr7_+_34794829 | 0.95 |
ENSDART00000009698
ENSDART00000075089 ENSDART00000173456 |
esrp2
|
epithelial splicing regulatory protein 2 |
| chr1_+_1599979 | 0.95 |
ENSDART00000097626
|
urp2
|
urotensin II-related peptide |
| chr10_-_7858553 | 0.94 |
ENSDART00000182010
|
inpp5ja
|
inositol polyphosphate-5-phosphatase Ja |
| chr24_-_24271629 | 0.94 |
ENSDART00000135060
|
rps6ka3b
|
ribosomal protein S6 kinase, polypeptide 3b |
| chr13_-_856525 | 0.93 |
ENSDART00000143356
|
TMEM14A
|
zgc:163080 |
| chr10_+_44903676 | 0.92 |
ENSDART00000158553
|
zgc:114173
|
zgc:114173 |
| chr11_-_45152702 | 0.92 |
ENSDART00000168066
|
afmid
|
arylformamidase |
| chr15_+_26941063 | 0.92 |
ENSDART00000149957
|
bcas3
|
breast carcinoma amplified sequence 3 |
| chr7_-_31794476 | 0.90 |
ENSDART00000142385
|
nap1l4b
|
nucleosome assembly protein 1-like 4b |
| chr22_+_1285182 | 0.90 |
ENSDART00000171750
|
si:ch73-138e16.3
|
si:ch73-138e16.3 |
| chr6_+_2951645 | 0.90 |
ENSDART00000183181
ENSDART00000181271 |
ptprfa
|
protein tyrosine phosphatase, receptor type, f, a |
| chr25_+_10458990 | 0.88 |
ENSDART00000130354
ENSDART00000044738 |
ric8a
|
RIC8 guanine nucleotide exchange factor A |
| chr12_+_25432627 | 0.87 |
ENSDART00000011662
|
ppm1bb
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Bb |
| chr3_-_1146497 | 0.84 |
ENSDART00000149061
|
si:ch73-211l13.2
|
si:ch73-211l13.2 |
| chr3_-_5964557 | 0.83 |
ENSDART00000184738
|
BX284638.1
|
|
| chr18_-_18942098 | 0.80 |
ENSDART00000100458
|
si:dkey-73n10.1
|
si:dkey-73n10.1 |
| chr7_+_57088920 | 0.79 |
ENSDART00000024076
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr16_+_49647402 | 0.79 |
ENSDART00000015694
ENSDART00000132547 |
rab5ab
|
RAB5A, member RAS oncogene family, b |
| chr6_-_6448519 | 0.75 |
ENSDART00000180157
ENSDART00000191112 |
si:ch211-194e18.2
|
si:ch211-194e18.2 |
| chr20_+_37866171 | 0.74 |
ENSDART00000153190
|
vash2
|
vasohibin 2 |
| chr17_+_35097024 | 0.74 |
ENSDART00000026152
|
asap2a
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2a |
| chr6_+_37752781 | 0.73 |
ENSDART00000154364
|
herc2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr25_-_35542739 | 0.72 |
ENSDART00000097651
|
si:ch211-87j1.4
|
si:ch211-87j1.4 |
| chr3_+_17878466 | 0.72 |
ENSDART00000180218
|
dnajc7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr19_-_27334394 | 0.72 |
ENSDART00000052359
|
gtf2h4
|
general transcription factor IIH, polypeptide 4 |
| chr1_-_30723241 | 0.71 |
ENSDART00000152175
ENSDART00000152150 |
MZT1
|
si:dkey-15d12.2 |
| chr3_+_17878124 | 0.70 |
ENSDART00000166430
ENSDART00000163421 ENSDART00000121473 |
dnajc7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr7_-_69636502 | 0.69 |
ENSDART00000126739
|
tspan5a
|
tetraspanin 5a |
Network of associatons between targets according to the STRING database.
First level regulatory network of ybx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.0 | GO:0010142 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 2.6 | 7.9 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 2.6 | 7.8 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 1.2 | 3.7 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 1.1 | 3.3 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 1.0 | 5.2 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 1.0 | 4.1 | GO:0034164 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 1.0 | 3.0 | GO:0050428 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.9 | 19.8 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.9 | 3.7 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.9 | 18.8 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.8 | 4.6 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.7 | 7.4 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.7 | 12.0 | GO:0043687 | protein N-linked glycosylation via asparagine(GO:0018279) post-translational protein modification(GO:0043687) |
| 0.7 | 2.7 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.6 | 6.1 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.6 | 3.0 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.6 | 2.3 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.5 | 2.0 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.5 | 3.8 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.5 | 10.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.4 | 1.8 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.4 | 1.3 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.4 | 16.5 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.4 | 3.7 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.4 | 2.0 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.4 | 15.8 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.4 | 2.0 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.3 | 4.9 | GO:0034033 | nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.3 | 4.0 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.3 | 1.0 | GO:1990868 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.3 | 2.5 | GO:0055129 | arginine catabolic process(GO:0006527) proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.3 | 2.8 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.3 | 4.1 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.3 | 2.4 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.3 | 0.8 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.3 | 2.6 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.3 | 1.0 | GO:0010898 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.3 | 1.3 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.2 | 2.9 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.2 | 6.3 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.2 | 4.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 3.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.2 | 2.1 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.2 | 2.1 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.2 | 1.9 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.2 | 1.2 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) negative regulation of intracellular protein transport(GO:0090317) |
| 0.2 | 2.8 | GO:0007032 | endosome organization(GO:0007032) |
| 0.2 | 1.0 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.2 | 6.1 | GO:0072350 | citrate metabolic process(GO:0006101) tricarboxylic acid metabolic process(GO:0072350) |
| 0.2 | 1.3 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.1 | 5.9 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 1.6 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 2.4 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 0.7 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 1.8 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 0.9 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.1 | 0.9 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 1.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 1.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 8.7 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 1.5 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.1 | 1.1 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 0.9 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 5.9 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.1 | 2.9 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.1 | 10.1 | GO:0006457 | protein folding(GO:0006457) |
| 0.1 | 1.1 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 1.0 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 1.6 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.1 | 0.3 | GO:0060760 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.1 | 1.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 1.7 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.1 | 5.2 | GO:0008202 | steroid metabolic process(GO:0008202) |
| 0.0 | 1.1 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.7 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 4.7 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 1.0 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 2.8 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.9 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 4.9 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.3 | GO:0051299 | mitotic centrosome separation(GO:0007100) centrosome separation(GO:0051299) |
| 0.0 | 0.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.4 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 5.2 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 4.3 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.5 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.0 | 1.8 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.5 | GO:0043489 | RNA stabilization(GO:0043489) |
| 0.0 | 1.6 | GO:0009306 | protein secretion(GO:0009306) |
| 0.0 | 1.8 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 0.3 | GO:0060324 | face development(GO:0060324) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.9 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.0 | 3.1 | GO:0006887 | exocytosis(GO:0006887) |
| 0.0 | 0.1 | GO:0090134 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.9 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.7 | 2.8 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.6 | 1.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.6 | 6.1 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.5 | 10.3 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.4 | 5.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.4 | 3.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 1.0 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.2 | 0.8 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.2 | 2.4 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.2 | 3.7 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.2 | 21.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 1.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 1.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 2.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 1.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 5.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.7 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 1.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.7 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 0.5 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 0.5 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 1.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 3.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 10.5 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 40.9 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 3.8 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 7.2 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 1.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 2.1 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 3.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 6.3 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 3.3 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.7 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 3.8 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.1 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.0 | 1.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 1.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.5 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 8.2 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 1.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.8 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.9 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 1.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 18.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.5 | GO:0005681 | spliceosomal complex(GO:0005681) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 12.0 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 2.7 | 8.0 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 1.7 | 5.2 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 1.3 | 19.8 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 1.3 | 9.1 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 1.1 | 4.6 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 1.0 | 3.0 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.9 | 4.7 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.9 | 18.8 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.9 | 5.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.8 | 12.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.8 | 3.0 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.6 | 3.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.5 | 3.6 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.5 | 3.8 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.5 | 5.5 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.4 | 1.3 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.4 | 5.7 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.4 | 2.8 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.4 | 2.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.4 | 2.8 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.3 | 6.1 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.3 | 3.7 | GO:0008556 | potassium-transporting ATPase activity(GO:0008556) |
| 0.3 | 1.3 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.3 | 0.9 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.3 | 1.5 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.3 | 2.9 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.3 | 2.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.2 | 1.0 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.2 | 6.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.2 | 1.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.2 | 1.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 11.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.2 | 1.6 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.2 | 2.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.2 | 8.5 | GO:0071949 | FAD binding(GO:0071949) |
| 0.2 | 5.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 3.7 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 1.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 1.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 1.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.9 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.1 | 4.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 1.8 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 2.5 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 1.3 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 0.9 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 2.0 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 3.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 0.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.5 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 2.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 1.1 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 1.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 1.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 6.7 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 18.9 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 3.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 1.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 2.4 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 4.9 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 2.4 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 1.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 1.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 11.8 | GO:0016791 | phosphatase activity(GO:0016791) |
| 0.0 | 7.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 7.2 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 2.0 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 1.7 | GO:0043492 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 3.0 | GO:0000989 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 7.5 | GO:0022803 | channel activity(GO:0015267) passive transmembrane transporter activity(GO:0022803) |
| 0.0 | 1.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 19.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.3 | 4.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 5.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 8.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 1.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 4.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.2 | 2.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 1.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 1.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.1 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 34.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 1.4 | 34.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 1.1 | 19.8 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.7 | 3.0 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.5 | 4.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.4 | 8.6 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.4 | 5.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.4 | 4.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.3 | 2.0 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.3 | 3.6 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.3 | 2.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 1.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 3.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.2 | 3.1 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.2 | 2.8 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 1.8 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 1.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 2.7 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 1.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 5.6 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.1 | 0.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 2.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.0 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.7 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 2.0 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 1.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.0 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |