Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
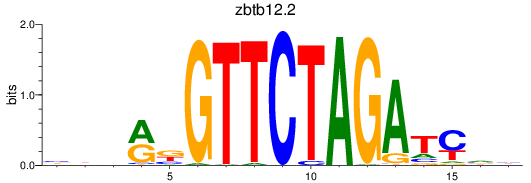
Results for zbtb12.2
Z-value: 0.59
Transcription factors associated with zbtb12.2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zbtb12.2
|
ENSDARG00000070658 | zinc finger and BTB domain containing 12, tandem duplicate 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zbtb12.2 | dr11_v1_chr19_-_26964348_26964348 | 0.37 | 2.0e-04 | Click! |
Activity profile of zbtb12.2 motif
Sorted Z-values of zbtb12.2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_26522760 | 5.98 |
ENSDART00000142417
ENSDART00000135606 ENSDART00000122668 |
si:dkey-205h13.1
|
si:dkey-205h13.1 |
| chr24_-_28259127 | 5.54 |
ENSDART00000149589
|
prkag2a
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit a |
| chr23_-_26521970 | 4.91 |
ENSDART00000143712
|
si:dkey-205h13.1
|
si:dkey-205h13.1 |
| chr17_+_53425037 | 3.16 |
ENSDART00000154983
|
fabp10b
|
fatty acid binding protein 10b, liver basic |
| chr21_-_13672195 | 2.84 |
ENSDART00000146717
|
clic3
|
chloride intracellular channel 3 |
| chr5_+_25311309 | 2.63 |
ENSDART00000169638
|
wu:fa19b12
|
wu:fa19b12 |
| chr10_-_15048781 | 2.41 |
ENSDART00000038401
ENSDART00000155674 |
si:ch211-95j8.2
|
si:ch211-95j8.2 |
| chr12_-_26430507 | 2.35 |
ENSDART00000153214
|
synpo2lb
|
synaptopodin 2-like b |
| chr3_-_2033105 | 2.25 |
ENSDART00000135249
|
si:dkey-88j15.3
|
si:dkey-88j15.3 |
| chr13_-_45475289 | 2.24 |
ENSDART00000043345
|
rsrp1
|
arginine/serine-rich protein 1 |
| chr17_+_53424415 | 1.79 |
ENSDART00000157022
|
slc9a1b
|
solute carrier family 9 member A1b |
| chr20_-_29532939 | 1.75 |
ENSDART00000049224
ENSDART00000062377 |
taf1b
|
TATA box binding protein (Tbp)-associated factor, RNA polymerase I, B |
| chr16_+_53526135 | 1.72 |
ENSDART00000083558
|
smpd5
|
sphingomyelin phosphodiesterase 5 |
| chr6_-_54126463 | 1.62 |
ENSDART00000161059
|
tusc2a
|
tumor suppressor candidate 2a |
| chr7_-_38659477 | 1.59 |
ENSDART00000138071
|
npsn
|
nephrosin |
| chr3_-_31784082 | 1.49 |
ENSDART00000134201
|
kcnh6a
|
potassium voltage-gated channel, subfamily H (eag-related), member 6a |
| chr18_+_8402129 | 1.38 |
ENSDART00000081154
ENSDART00000171974 |
prpf18
|
PRP18 pre-mRNA processing factor 18 homolog (yeast) |
| chr2_+_22602301 | 1.37 |
ENSDART00000038514
|
sept2
|
septin 2 |
| chr3_-_31783737 | 1.36 |
ENSDART00000090809
|
kcnh6a
|
potassium voltage-gated channel, subfamily H (eag-related), member 6a |
| chr20_-_44496245 | 1.35 |
ENSDART00000012229
|
fkbp1b
|
FK506 binding protein 1b |
| chr10_+_29849497 | 1.23 |
ENSDART00000099994
ENSDART00000132212 |
hspa8
|
heat shock protein 8 |
| chr24_+_42148140 | 1.17 |
ENSDART00000010658
|
serpinb1l3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 3 |
| chr19_+_20793388 | 1.15 |
ENSDART00000142463
|
txnl4a
|
thioredoxin-like 4A |
| chr6_-_2154137 | 1.15 |
ENSDART00000162656
|
tgm5l
|
transglutaminase 5, like |
| chr16_-_46619967 | 1.12 |
ENSDART00000158341
|
tmem176l.3a
|
transmembrane protein 176l.3a |
| chr19_+_20793237 | 1.12 |
ENSDART00000014774
|
txnl4a
|
thioredoxin-like 4A |
| chr22_+_2239974 | 1.11 |
ENSDART00000141993
|
znf1144
|
zinc finger protein 1144 |
| chr22_+_9862243 | 1.10 |
ENSDART00000105942
|
si:dkey-253d23.3
|
si:dkey-253d23.3 |
| chr11_-_18254 | 1.10 |
ENSDART00000167814
|
prr13
|
proline rich 13 |
| chr11_-_18449 | 1.09 |
ENSDART00000172050
|
prr13
|
proline rich 13 |
| chr3_-_61345723 | 1.01 |
ENSDART00000156749
|
si:dkey-111k8.5
|
si:dkey-111k8.5 |
| chr6_-_22369125 | 0.99 |
ENSDART00000083038
|
nprl2
|
NPR2 like, GATOR1 complex subunit |
| chr19_-_24136233 | 0.98 |
ENSDART00000143365
|
thap7
|
THAP domain containing 7 |
| chr21_-_28737320 | 0.97 |
ENSDART00000098696
ENSDART00000124826 ENSDART00000125652 |
nrg2a
|
neuregulin 2a |
| chr5_-_29570141 | 0.94 |
ENSDART00000043259
|
entpd2a.2
|
ectonucleoside triphosphate diphosphohydrolase 2a, tandem duplicate 2 |
| chr1_+_218524 | 0.94 |
ENSDART00000109529
|
tmco3
|
transmembrane and coiled-coil domains 3 |
| chr14_+_46342882 | 0.86 |
ENSDART00000193707
ENSDART00000060577 |
tmem33
|
transmembrane protein 33 |
| chr19_+_8985230 | 0.86 |
ENSDART00000018973
|
scamp3
|
secretory carrier membrane protein 3 |
| chr6_+_20954400 | 0.85 |
ENSDART00000143248
ENSDART00000165806 |
stk11ip
|
serine/threonine kinase 11 interacting protein |
| chr16_-_48430272 | 0.83 |
ENSDART00000005927
|
rad21a
|
RAD21 cohesin complex component a |
| chr8_+_24281512 | 0.82 |
ENSDART00000062845
|
mmp9
|
matrix metallopeptidase 9 |
| chr20_+_9355912 | 0.80 |
ENSDART00000142337
|
si:dkey-174m14.3
|
si:dkey-174m14.3 |
| chr6_-_24392426 | 0.80 |
ENSDART00000163965
|
brdt
|
bromodomain, testis-specific |
| chr16_+_25989943 | 0.80 |
ENSDART00000149113
|
si:ch73-103b2.1
|
si:ch73-103b2.1 |
| chr3_-_61354239 | 0.77 |
ENSDART00000154312
|
znf1134
|
zinc finger protein 1134 |
| chr3_+_62339264 | 0.76 |
ENSDART00000174569
|
BX470259.3
|
|
| chr22_+_2686673 | 0.75 |
ENSDART00000161580
|
si:dkey-20i20.15
|
si:dkey-20i20.15 |
| chr15_+_38299563 | 0.74 |
ENSDART00000099375
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr3_+_43374571 | 0.68 |
ENSDART00000182497
|
zfand2a
|
zinc finger, AN1-type domain 2A |
| chr6_+_12482599 | 0.68 |
ENSDART00000090316
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr17_-_32865788 | 0.67 |
ENSDART00000077476
|
prox1a
|
prospero homeobox 1a |
| chr19_+_26640096 | 0.66 |
ENSDART00000067793
|
ints3
|
integrator complex subunit 3 |
| chr7_+_29024282 | 0.65 |
ENSDART00000076425
|
cog4
|
component of oligomeric golgi complex 4 |
| chr19_+_3215466 | 0.65 |
ENSDART00000181288
|
zgc:86598
|
zgc:86598 |
| chr1_+_16127825 | 0.63 |
ENSDART00000122503
|
tusc3
|
tumor suppressor candidate 3 |
| chr12_+_9849476 | 0.62 |
ENSDART00000110458
|
fam117ab
|
family with sequence similarity 117, member Ab |
| chr6_+_11397269 | 0.61 |
ENSDART00000114260
|
senp2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr22_+_1540849 | 0.58 |
ENSDART00000192088
|
si:ch211-255f4.7
|
si:ch211-255f4.7 |
| chr22_+_1734981 | 0.57 |
ENSDART00000158195
|
znf1159
|
zinc finger protein 1159 |
| chr13_-_4223955 | 0.56 |
ENSDART00000113060
|
dnph1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr21_-_43171249 | 0.54 |
ENSDART00000021037
ENSDART00000148630 |
hspa4a
|
heat shock protein 4a |
| chr5_-_66823750 | 0.51 |
ENSDART00000041441
ENSDART00000112488 |
stip1
|
stress-induced phosphoprotein 1 |
| chr21_+_1587722 | 0.49 |
ENSDART00000013581
|
wdr91
|
WD repeat domain 91 |
| chr22_+_1837448 | 0.48 |
ENSDART00000160015
|
znf1183
|
zinc finger protein 1183 |
| chr19_-_25519310 | 0.48 |
ENSDART00000089882
|
C1GALT1 (1 of many)
|
si:dkey-202e17.1 |
| chr21_+_20386865 | 0.47 |
ENSDART00000144366
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr3_-_36364903 | 0.47 |
ENSDART00000028883
|
gna13b
|
guanine nucleotide binding protein (G protein), alpha 13b |
| chr16_+_38659475 | 0.47 |
ENSDART00000023238
|
emc2
|
ER membrane protein complex subunit 2 |
| chr19_+_3140313 | 0.46 |
ENSDART00000125504
|
zgc:86598
|
zgc:86598 |
| chr21_+_43171013 | 0.45 |
ENSDART00000151362
|
zcchc10
|
zinc finger, CCHC domain containing 10 |
| chr19_-_25519612 | 0.44 |
ENSDART00000133150
|
C1GALT1 (1 of many)
|
si:dkey-202e17.1 |
| chr3_+_43373867 | 0.42 |
ENSDART00000159455
ENSDART00000172425 |
zfand2a
|
zinc finger, AN1-type domain 2A |
| chr5_+_31944270 | 0.36 |
ENSDART00000147850
|
ungb
|
uracil DNA glycosylase b |
| chr17_-_12249990 | 0.36 |
ENSDART00000177889
ENSDART00000155545 |
ahctf1
|
AT hook containing transcription factor 1 |
| chr16_-_42175617 | 0.36 |
ENSDART00000084715
|
alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr3_-_15645405 | 0.36 |
ENSDART00000133957
|
si:dkey-93n13.1
|
si:dkey-93n13.1 |
| chr7_-_60156409 | 0.35 |
ENSDART00000006802
|
cct7
|
chaperonin containing TCP1, subunit 7 (eta) |
| chr1_-_59021796 | 0.34 |
ENSDART00000168314
|
zgc:153247
|
zgc:153247 |
| chr5_-_42950963 | 0.32 |
ENSDART00000149868
|
grsf1
|
G-rich RNA sequence binding factor 1 |
| chr7_+_34297271 | 0.32 |
ENSDART00000180342
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr22_-_14475927 | 0.32 |
ENSDART00000135768
|
lrp1ba
|
low density lipoprotein receptor-related protein 1Ba |
| chr1_+_54194801 | 0.31 |
ENSDART00000186802
|
CABZ01067150.1
|
|
| chr2_-_20981907 | 0.30 |
ENSDART00000113384
|
lyrm4
|
LYR motif containing 4 |
| chr22_+_2144278 | 0.30 |
ENSDART00000162173
ENSDART00000159914 ENSDART00000160192 |
znf1164
|
zinc finger protein 1164 |
| chr13_+_23623346 | 0.27 |
ENSDART00000057619
|
il22ra2
|
interleukin 22 receptor, alpha 2 |
| chr16_-_10837245 | 0.27 |
ENSDART00000036891
|
rabac1
|
Rab acceptor 1 (prenylated) |
| chr23_+_34005792 | 0.25 |
ENSDART00000132668
|
si:ch211-207e14.4
|
si:ch211-207e14.4 |
| chr7_+_59649399 | 0.23 |
ENSDART00000123520
ENSDART00000040771 |
rpl34
|
ribosomal protein L34 |
| chr22_+_1300587 | 0.21 |
ENSDART00000124161
|
si:ch73-138e16.5
|
si:ch73-138e16.5 |
| chr17_-_50436199 | 0.21 |
ENSDART00000156494
|
si:ch211-235i11.6
|
si:ch211-235i11.6 |
| chr17_-_50454957 | 0.19 |
ENSDART00000171616
ENSDART00000075195 |
si:ch211-235i11.7
|
si:ch211-235i11.7 |
| chr22_+_2512154 | 0.18 |
ENSDART00000097363
|
zgc:173726
|
zgc:173726 |
| chr22_+_9287929 | 0.14 |
ENSDART00000193522
|
si:ch211-250k18.7
|
si:ch211-250k18.7 |
| chr22_+_2048566 | 0.13 |
ENSDART00000169234
|
si:dkey-1b17.9
|
si:dkey-1b17.9 |
| chr24_-_7957581 | 0.12 |
ENSDART00000145815
|
txndc5
|
thioredoxin domain containing 5 |
| chr10_-_17284055 | 0.11 |
ENSDART00000167464
|
GNAZ
|
G protein subunit alpha z |
| chr22_+_9239831 | 0.11 |
ENSDART00000133720
|
si:ch211-250k18.5
|
si:ch211-250k18.5 |
| chr7_+_21768452 | 0.10 |
ENSDART00000127719
|
kdm6ba
|
lysine (K)-specific demethylase 6B, a |
| chr20_-_38449903 | 0.10 |
ENSDART00000136303
|
itpkb
|
inositol-trisphosphate 3-kinase B |
| chr4_+_20085114 | 0.09 |
ENSDART00000186698
ENSDART00000188635 |
ppp6r2a
|
protein phosphatase 6, regulatory subunit 2a |
| chr16_-_45398408 | 0.09 |
ENSDART00000004052
|
rgl2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr18_+_28106139 | 0.09 |
ENSDART00000089615
|
kiaa1549lb
|
KIAA1549-like b |
| chr3_-_136043 | 0.09 |
ENSDART00000160266
|
zgc:110249
|
zgc:110249 |
| chr23_-_38054 | 0.07 |
ENSDART00000170393
|
CABZ01074076.1
|
|
| chr3_-_7129057 | 0.07 |
ENSDART00000125947
|
BX005085.1
|
|
| chr9_-_180334 | 0.03 |
ENSDART00000180339
|
zgc:158619
|
zgc:158619 |
| chr1_+_32521469 | 0.02 |
ENSDART00000113818
ENSDART00000152580 |
nlgn4a
|
neuroligin 4a |
| chr1_-_40341306 | 0.02 |
ENSDART00000190649
|
maml3
|
mastermind-like transcriptional coactivator 3 |
| chr22_+_9862466 | 0.00 |
ENSDART00000146864
|
si:dkey-253d23.3
|
si:dkey-253d23.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of zbtb12.2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0099623 | regulation of membrane repolarization(GO:0060306) regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) regulation of cardiac muscle cell membrane repolarization(GO:0099623) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.5 | 1.4 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.3 | 1.7 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.3 | 1.6 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.3 | 0.8 | GO:0002676 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.3 | 0.8 | GO:0001207 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.2 | 1.2 | GO:1902946 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.2 | 0.6 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.2 | 1.7 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 0.8 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.2 | 1.8 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 0.9 | GO:0071405 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.1 | 5.5 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 1.0 | GO:0060347 | ventricular trabecula myocardium morphogenesis(GO:0003222) trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) skin epidermis development(GO:0098773) |
| 0.1 | 0.5 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 0.9 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.1 | 2.3 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 10.9 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.1 | 0.4 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.1 | 0.4 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.5 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.1 | 1.0 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 1.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.8 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 1.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.6 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.5 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.7 | GO:0044818 | mitotic G2/M transition checkpoint(GO:0044818) |
| 0.0 | 0.1 | GO:0061384 | trabecula morphogenesis(GO:0061383) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.7 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 2.8 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 1.1 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.4 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.7 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.3 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.5 | 5.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 3.6 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 1.0 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.8 | GO:0000798 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.7 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 1.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 0.5 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 1.4 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 0.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.7 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 2.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 1.7 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 2.3 | GO:0030018 | Z disc(GO:0030018) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0001163 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.4 | 10.9 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.2 | 5.5 | GO:0016208 | AMP binding(GO:0016208) |
| 0.2 | 1.8 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 0.5 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 1.7 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.4 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.1 | 0.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 1.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.9 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 0.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.9 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.5 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.6 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.9 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 2.8 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 1.3 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 2.8 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 2.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.5 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.4 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.7 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 2.3 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |