Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
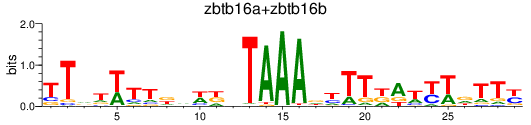
Results for zbtb16a+zbtb16b
Z-value: 0.95
Transcription factors associated with zbtb16a+zbtb16b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zbtb16a
|
ENSDARG00000007184 | zinc finger and BTB domain containing 16a |
|
zbtb16b
|
ENSDARG00000074526 | zinc finger and BTB domain containing 16b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zbtb16b | dr11_v1_chr15_-_18361475_18361475 | 0.23 | 2.4e-02 | Click! |
| zbtb16a | dr11_v1_chr21_-_23307653_23307796 | 0.06 | 5.4e-01 | Click! |
Activity profile of zbtb16a+zbtb16b motif
Sorted Z-values of zbtb16a+zbtb16b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_31270761 | 21.20 |
ENSDART00000176187
|
apobb.1
|
apolipoprotein Bb, tandem duplicate 1 |
| chr16_-_2414063 | 15.91 |
ENSDART00000073621
|
zgc:152945
|
zgc:152945 |
| chr13_+_17672527 | 13.20 |
ENSDART00000148269
ENSDART00000137776 |
comtd1
|
catechol-O-methyltransferase domain containing 1 |
| chr2_-_1514001 | 12.90 |
ENSDART00000057736
|
c8b
|
complement component 8, beta polypeptide |
| chr2_+_37875789 | 12.48 |
ENSDART00000036318
ENSDART00000127679 |
cbln13
|
cerebellin 13 |
| chr3_-_45848043 | 11.55 |
ENSDART00000055132
|
igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr8_+_39724138 | 11.10 |
ENSDART00000009323
|
pla2g1b
|
phospholipase A2, group IB (pancreas) |
| chr3_-_45848257 | 11.09 |
ENSDART00000147198
|
igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr17_+_33340675 | 9.55 |
ENSDART00000184396
ENSDART00000077553 |
xdh
|
xanthine dehydrogenase |
| chr11_+_13224281 | 7.81 |
ENSDART00000102557
ENSDART00000178706 |
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr24_-_33578392 | 7.55 |
ENSDART00000079476
|
LO018309.1
|
|
| chr14_-_21123551 | 6.69 |
ENSDART00000171679
ENSDART00000165882 |
si:dkey-74k8.4
|
si:dkey-74k8.4 |
| chr25_-_17378881 | 6.18 |
ENSDART00000064586
|
cyp2x7
|
cytochrome P450, family 2, subfamily X, polypeptide 7 |
| chr4_+_2230701 | 6.02 |
ENSDART00000080439
|
cmah
|
cytidine monophospho-N-acetylneuraminic acid hydroxylase |
| chr21_+_51521 | 5.14 |
ENSDART00000007672
|
dmgdh
|
dimethylglycine dehydrogenase |
| chr4_-_76270779 | 4.74 |
ENSDART00000183709
ENSDART00000192689 |
si:ch211-106j21.4
|
si:ch211-106j21.4 |
| chr4_-_12723585 | 4.66 |
ENSDART00000185639
|
mgst1.2
|
microsomal glutathione S-transferase 1.2 |
| chr4_-_75997108 | 4.32 |
ENSDART00000163113
|
si:ch211-232d10.1
|
si:ch211-232d10.1 |
| chr9_-_34882516 | 4.20 |
ENSDART00000011163
|
asmtl
|
acetylserotonin O-methyltransferase-like |
| chr18_-_34171280 | 4.18 |
ENSDART00000122321
|
slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr20_+_26880668 | 4.10 |
ENSDART00000077769
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr14_-_14659023 | 3.96 |
ENSDART00000170355
ENSDART00000159888 ENSDART00000172241 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr2_-_23931536 | 3.81 |
ENSDART00000121885
|
tgfbr1a
|
transforming growth factor, beta receptor 1 a |
| chr20_-_5229157 | 3.74 |
ENSDART00000114851
ENSDART00000182876 |
ccdc85cb
|
coiled-coil domain containing 85C, b |
| chr21_+_10712823 | 3.73 |
ENSDART00000123476
|
lman1
|
lectin, mannose-binding, 1 |
| chr3_+_62161184 | 3.67 |
ENSDART00000090370
ENSDART00000192665 |
noxo1a
|
NADPH oxidase organizer 1a |
| chr18_-_34170918 | 3.64 |
ENSDART00000015079
|
slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr18_-_34549721 | 3.60 |
ENSDART00000137101
ENSDART00000021880 |
ssr3
|
signal sequence receptor, gamma |
| chr4_-_75686458 | 3.55 |
ENSDART00000172032
|
si:dkey-165o8.1
|
si:dkey-165o8.1 |
| chr7_+_61184104 | 3.44 |
ENSDART00000110671
|
zgc:194930
|
zgc:194930 |
| chr17_+_16564921 | 3.29 |
ENSDART00000151904
|
foxn3
|
forkhead box N3 |
| chr2_-_55298075 | 3.17 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr10_-_7666810 | 3.16 |
ENSDART00000191479
|
pcyox1
|
prenylcysteine oxidase 1 |
| chr4_-_17725008 | 2.93 |
ENSDART00000016658
|
chpt1
|
choline phosphotransferase 1 |
| chr21_-_11970199 | 2.89 |
ENSDART00000114524
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr12_+_20587179 | 2.80 |
ENSDART00000170127
|
arsg
|
arylsulfatase G |
| chr3_-_50136424 | 2.77 |
ENSDART00000188843
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr20_+_36628059 | 2.68 |
ENSDART00000062898
|
ephx1
|
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr18_+_38755023 | 2.67 |
ENSDART00000010177
ENSDART00000193136 |
onecut1
|
one cut homeobox 1 |
| chr8_-_29713595 | 2.67 |
ENSDART00000131988
ENSDART00000077637 |
mpeg1.1
|
macrophage expressed 1, tandem duplicate 1 |
| chr12_-_45197038 | 2.67 |
ENSDART00000016635
|
bccip
|
BRCA2 and CDKN1A interacting protein |
| chr15_-_17960228 | 2.49 |
ENSDART00000155898
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr4_+_10003523 | 2.30 |
ENSDART00000148105
|
naf1
|
nuclear assembly factor 1 homolog (S. cerevisiae) |
| chr5_+_66250856 | 2.24 |
ENSDART00000132789
|
secisbp2
|
SECIS binding protein 2 |
| chr12_+_10448226 | 2.22 |
ENSDART00000152824
|
crym
|
crystallin, mu |
| chr19_+_43753995 | 2.12 |
ENSDART00000058504
|
SLC17A3
|
si:ch1073-513e17.1 |
| chr22_+_10159512 | 2.10 |
ENSDART00000184415
|
rpp14
|
ribonuclease P/MRP 14 subunit |
| chr3_+_18807006 | 2.07 |
ENSDART00000180091
|
tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr9_+_50316921 | 2.03 |
ENSDART00000098687
|
GRB14
|
growth factor receptor bound protein 14 |
| chr20_+_17739923 | 1.95 |
ENSDART00000024627
|
cdh2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr17_+_16565185 | 1.94 |
ENSDART00000136874
|
foxn3
|
forkhead box N3 |
| chr8_-_6825588 | 1.91 |
ENSDART00000135834
|
dock5
|
dedicator of cytokinesis 5 |
| chr1_+_44173245 | 1.89 |
ENSDART00000159450
ENSDART00000106048 ENSDART00000157763 |
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr12_-_33746111 | 1.87 |
ENSDART00000127203
|
llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr22_-_31020690 | 1.84 |
ENSDART00000130604
|
ssuh2.4
|
ssu-2 homolog, tandem duplicate 4 |
| chr13_-_37474989 | 1.83 |
ENSDART00000114136
|
wdr89
|
WD repeat domain 89 |
| chr18_-_42071876 | 1.81 |
ENSDART00000193288
ENSDART00000189021 ENSDART00000019245 |
arhgap42b
|
Rho GTPase activating protein 42b |
| chr17_+_39242437 | 1.76 |
ENSDART00000156138
ENSDART00000128863 |
zgc:174356
|
zgc:174356 |
| chr15_-_25094026 | 1.73 |
ENSDART00000129154
|
exo5
|
exonuclease 5 |
| chr21_-_32462856 | 1.73 |
ENSDART00000147318
|
zgc:123105
|
zgc:123105 |
| chr6_+_7553085 | 1.73 |
ENSDART00000150939
ENSDART00000151114 |
myh10
|
myosin, heavy chain 10, non-muscle |
| chr17_-_23603263 | 1.71 |
ENSDART00000079559
|
ifit16
|
interferon-induced protein with tetratricopeptide repeats 16 |
| chr17_+_41343167 | 1.67 |
ENSDART00000156405
|
mocs1
|
molybdenum cofactor synthesis 1 |
| chr21_-_43636595 | 1.61 |
ENSDART00000151115
ENSDART00000151486 ENSDART00000151778 |
si:ch1073-263o8.2
|
si:ch1073-263o8.2 |
| chr22_+_1170294 | 1.60 |
ENSDART00000159761
ENSDART00000169809 |
irf6
|
interferon regulatory factor 6 |
| chr22_-_16403602 | 1.59 |
ENSDART00000183417
|
lama3
|
laminin, alpha 3 |
| chr4_+_5868034 | 1.58 |
ENSDART00000166591
|
utp20
|
UTP20 small subunit (SSU) processome component |
| chr10_-_7636340 | 1.53 |
ENSDART00000166234
|
ubxn8
|
UBX domain protein 8 |
| chr8_+_19624589 | 1.52 |
ENSDART00000185698
|
slc35a3a
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3a |
| chr11_-_21404358 | 1.48 |
ENSDART00000129062
|
ikbke
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr6_+_54221654 | 1.48 |
ENSDART00000128456
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr19_-_4137087 | 1.45 |
ENSDART00000164611
|
rragcb
|
Ras-related GTP binding Cb |
| chr22_-_6194517 | 1.44 |
ENSDART00000134757
ENSDART00000181598 ENSDART00000129829 |
si:rp71-36a1.5
|
si:rp71-36a1.5 |
| chr23_+_32406009 | 1.42 |
ENSDART00000155793
|
si:ch211-66i15.5
|
si:ch211-66i15.5 |
| chr7_-_51710015 | 1.42 |
ENSDART00000009184
|
hdac8
|
histone deacetylase 8 |
| chr22_-_5252005 | 1.40 |
ENSDART00000132942
ENSDART00000081801 |
ncln
|
nicalin |
| chr10_+_29137482 | 1.40 |
ENSDART00000178280
|
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr19_+_1097393 | 1.38 |
ENSDART00000168218
|
CABZ01111953.1
|
|
| chr25_+_21716263 | 1.32 |
ENSDART00000148920
|
znf277
|
zinc finger protein 277 |
| chr13_-_5978433 | 1.32 |
ENSDART00000102555
|
actr2b
|
ARP2 actin related protein 2b homolog |
| chr6_-_30689126 | 1.29 |
ENSDART00000065211
|
pars2
|
prolyl-tRNA synthetase 2, mitochondrial |
| chr23_+_2666944 | 1.27 |
ENSDART00000192861
|
CABZ01057928.1
|
|
| chr2_-_41124013 | 1.27 |
ENSDART00000134756
|
hs6st1a
|
heparan sulfate 6-O-sulfotransferase 1a |
| chr23_-_15916316 | 1.25 |
ENSDART00000134096
ENSDART00000042469 ENSDART00000146605 |
mrgbp
|
MRG/MORF4L binding protein |
| chr20_-_7080427 | 1.24 |
ENSDART00000140166
ENSDART00000023677 |
si:ch211-121a2.2
|
si:ch211-121a2.2 |
| chr22_+_24646339 | 1.24 |
ENSDART00000183865
|
lpar3
|
lysophosphatidic acid receptor 3 |
| chr12_+_28854963 | 1.20 |
ENSDART00000153227
|
nfe2l1b
|
nuclear factor, erythroid 2-like 1b |
| chr12_+_2870671 | 1.19 |
ENSDART00000165225
|
prkar1ab
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) b |
| chr19_-_27588842 | 1.19 |
ENSDART00000121643
|
si:dkeyp-46h3.2
|
si:dkeyp-46h3.2 |
| chr17_-_13058515 | 1.17 |
ENSDART00000172450
ENSDART00000170255 |
CU469462.1
|
|
| chr3_-_50954607 | 1.17 |
ENSDART00000163810
|
cdc42ep4a
|
CDC42 effector protein (Rho GTPase binding) 4a |
| chr14_-_32876280 | 1.16 |
ENSDART00000173168
|
si:rp71-46j2.7
|
si:rp71-46j2.7 |
| chr13_+_19283936 | 1.14 |
ENSDART00000111462
|
eif3s10
|
eukaryotic translation initiation factor 3, subunit 10 (theta) |
| chr6_+_6828167 | 1.13 |
ENSDART00000181284
|
si:ch211-85n16.4
|
si:ch211-85n16.4 |
| chr5_-_31035198 | 1.13 |
ENSDART00000086534
|
cyb5d2
|
cytochrome b5 domain containing 2 |
| chr7_-_24838367 | 1.12 |
ENSDART00000139455
ENSDART00000012483 ENSDART00000131530 |
fam113
|
family with sequence similarity 113 |
| chr12_+_33460794 | 1.07 |
ENSDART00000007053
ENSDART00000142716 |
narf
|
nuclear prelamin A recognition factor |
| chr7_+_17947217 | 1.06 |
ENSDART00000101601
|
cth1
|
cysteine three histidine 1 |
| chr23_+_32406461 | 1.03 |
ENSDART00000179878
|
si:ch211-66i15.5
|
si:ch211-66i15.5 |
| chr1_-_52447364 | 1.01 |
ENSDART00000140740
|
si:ch211-217k17.10
|
si:ch211-217k17.10 |
| chr5_+_20570573 | 0.99 |
ENSDART00000137019
|
cmklr1
|
chemokine-like receptor 1 |
| chr12_+_3871452 | 0.98 |
ENSDART00000066546
|
nif3l1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr18_+_15841449 | 0.98 |
ENSDART00000141800
ENSDART00000091349 |
eea1
|
early endosome antigen 1 |
| chr25_+_10793478 | 0.97 |
ENSDART00000058339
ENSDART00000134923 |
ap3s2
|
adaptor-related protein complex 3, sigma 2 subunit |
| chr16_+_20947439 | 0.96 |
ENSDART00000137344
|
skap2
|
src kinase associated phosphoprotein 2 |
| chr19_-_425145 | 0.96 |
ENSDART00000164905
|
dus3l
|
dihydrouridine synthase 3-like (S. cerevisiae) |
| chr18_-_2639351 | 0.94 |
ENSDART00000168106
|
relt
|
RELT, TNF receptor |
| chr3_-_26190804 | 0.93 |
ENSDART00000136001
|
ypel3
|
yippee-like 3 |
| chr17_-_24714837 | 0.93 |
ENSDART00000154871
|
si:ch211-15d5.11
|
si:ch211-15d5.11 |
| chr1_+_49955869 | 0.92 |
ENSDART00000150517
|
gstcd
|
glutathione S-transferase, C-terminal domain containing |
| chr17_-_46990003 | 0.92 |
ENSDART00000155257
|
si:ch211-244k5.1
|
si:ch211-244k5.1 |
| chr21_-_35082715 | 0.92 |
ENSDART00000146454
|
adrb2b
|
adrenoceptor beta 2, surface b |
| chr12_+_48841419 | 0.90 |
ENSDART00000125331
|
dlg5b.1
|
discs, large homolog 5b (Drosophila), tandem duplicate 1 |
| chr14_-_9085349 | 0.90 |
ENSDART00000054710
|
polr1d
|
polymerase (RNA) I polypeptide D |
| chr8_+_26083808 | 0.89 |
ENSDART00000099283
|
dalrd3
|
DALR anticodon binding domain containing 3 |
| chr12_+_48841182 | 0.87 |
ENSDART00000109315
ENSDART00000185609 ENSDART00000187217 |
dlg5b.1
|
discs, large homolog 5b (Drosophila), tandem duplicate 1 |
| chr7_-_24838857 | 0.86 |
ENSDART00000179766
ENSDART00000180892 |
fam113
|
family with sequence similarity 113 |
| chr11_-_27917730 | 0.85 |
ENSDART00000173219
|
eif4g3a
|
eukaryotic translation initiation factor 4 gamma, 3a |
| chr22_+_29204885 | 0.84 |
ENSDART00000133409
ENSDART00000059820 |
ncf4
|
neutrophil cytosolic factor 4 |
| chr10_-_8955413 | 0.81 |
ENSDART00000110628
|
mocs2
|
molybdenum cofactor synthesis 2 |
| chr15_-_44663712 | 0.81 |
ENSDART00000192124
|
CABZ01061894.1
|
|
| chr18_-_50766660 | 0.80 |
ENSDART00000170663
ENSDART00000168601 |
zgc:158464
|
zgc:158464 |
| chr12_+_46512881 | 0.80 |
ENSDART00000105454
|
CU855711.1
|
|
| chr10_+_44956660 | 0.78 |
ENSDART00000169225
ENSDART00000189298 |
il1b
|
interleukin 1, beta |
| chr14_+_15620640 | 0.77 |
ENSDART00000188867
|
si:dkey-203a12.9
|
si:dkey-203a12.9 |
| chr15_-_25093680 | 0.77 |
ENSDART00000062695
|
exo5
|
exonuclease 5 |
| chr13_+_20007366 | 0.75 |
ENSDART00000147217
|
atrnl1a
|
attractin-like 1a |
| chr4_-_1839192 | 0.75 |
ENSDART00000003790
|
pwp1
|
PWP1 homolog (S. cerevisiae) |
| chr5_+_38726854 | 0.74 |
ENSDART00000138484
ENSDART00000142867 |
si:dkey-58f10.7
|
si:dkey-58f10.7 |
| chr9_+_45227028 | 0.73 |
ENSDART00000185579
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr14_+_3507326 | 0.73 |
ENSDART00000159326
|
gstp1
|
glutathione S-transferase pi 1 |
| chr16_+_28994709 | 0.73 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr3_-_13509283 | 0.73 |
ENSDART00000166832
|
slc25a19
|
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
| chr21_-_9383974 | 0.73 |
ENSDART00000160932
|
sdad1
|
SDA1 domain containing 1 |
| chr14_-_14705335 | 0.73 |
ENSDART00000157392
ENSDART00000159456 |
ogt.1
|
O-linked N-acetylglucosamine (GlcNAc) transferase, tandem duplicate 1 |
| chr21_-_41545995 | 0.71 |
ENSDART00000169977
|
bnip1b
|
BCL2 interacting protein 1b |
| chr24_-_40860603 | 0.71 |
ENSDART00000188032
|
CU633479.7
|
|
| chr6_+_15268685 | 0.71 |
ENSDART00000128090
ENSDART00000154417 |
ecrg4b
|
esophageal cancer related gene 4b |
| chr13_+_5978809 | 0.71 |
ENSDART00000102563
ENSDART00000121598 |
phf10
|
PHD finger protein 10 |
| chr6_+_55819038 | 0.70 |
ENSDART00000108786
|
si:ch211-81n22.1
|
si:ch211-81n22.1 |
| chr16_+_51159775 | 0.69 |
ENSDART00000161924
|
dhdds
|
dehydrodolichyl diphosphate synthase |
| chr17_-_12249990 | 0.69 |
ENSDART00000177889
ENSDART00000155545 |
ahctf1
|
AT hook containing transcription factor 1 |
| chr9_+_8894788 | 0.68 |
ENSDART00000132068
|
naxd
|
NAD(P)HX dehydratase |
| chr10_+_34315719 | 0.68 |
ENSDART00000135303
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr2_-_11027258 | 0.67 |
ENSDART00000081072
ENSDART00000193824 ENSDART00000187036 ENSDART00000097741 |
ssbp3a
|
single stranded DNA binding protein 3a |
| chr21_-_13051613 | 0.66 |
ENSDART00000190777
|
myorg
|
myogenesis regulating glycosidase (putative) |
| chr7_-_70665965 | 0.64 |
ENSDART00000097710
|
ppargc1a
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr4_-_64604842 | 0.64 |
ENSDART00000169862
|
znf1099
|
zinc finger protein 1099 |
| chr6_-_60049272 | 0.63 |
ENSDART00000057466
|
ccndbp1
|
cyclin D-type binding-protein 1 |
| chr16_-_45209684 | 0.63 |
ENSDART00000184595
|
fxyd1
|
FXYD domain containing ion transport regulator 1 (phospholemman) |
| chr16_+_32729223 | 0.62 |
ENSDART00000125663
|
si:dkey-165n16.1
|
si:dkey-165n16.1 |
| chr15_-_20190052 | 0.62 |
ENSDART00000157149
|
exoc3l2b
|
exocyst complex component 3-like 2b |
| chr5_-_71765780 | 0.61 |
ENSDART00000011955
|
gpsm1b
|
G protein signaling modulator 1b |
| chr6_+_3934738 | 0.60 |
ENSDART00000159673
|
dync1i2b
|
dynein, cytoplasmic 1, intermediate chain 2b |
| chr9_-_7378566 | 0.59 |
ENSDART00000144003
|
slc23a3
|
solute carrier family 23, member 3 |
| chr2_+_21048661 | 0.59 |
ENSDART00000156876
|
rreb1b
|
ras responsive element binding protein 1b |
| chr24_-_36175365 | 0.58 |
ENSDART00000065338
|
pak1ip1
|
PAK1 interacting protein 1 |
| chr17_-_10738001 | 0.58 |
ENSDART00000051526
|
jmjd7
|
jumonji domain containing 7 |
| chr15_-_25584888 | 0.56 |
ENSDART00000127571
|
si:dkey-54n8.2
|
si:dkey-54n8.2 |
| chr8_-_32354677 | 0.55 |
ENSDART00000138268
ENSDART00000133245 ENSDART00000179677 ENSDART00000174450 |
ipo11
|
importin 11 |
| chr14_+_49376011 | 0.55 |
ENSDART00000020961
|
tnip1
|
TNFAIP3 interacting protein 1 |
| chr17_+_16429826 | 0.53 |
ENSDART00000136078
|
efcab11
|
EF-hand calcium binding domain 11 |
| chr2_+_44512324 | 0.53 |
ENSDART00000155017
ENSDART00000156310 ENSDART00000156686 |
pask
|
PAS domain containing serine/threonine kinase |
| chr4_+_47656992 | 0.53 |
ENSDART00000161148
|
znf1040
|
zinc finger protein 1040 |
| chr20_-_24182689 | 0.53 |
ENSDART00000171184
|
map3k7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr10_+_36439293 | 0.53 |
ENSDART00000043802
|
uspl1
|
ubiquitin specific peptidase like 1 |
| chr4_-_1839352 | 0.52 |
ENSDART00000189215
|
pwp1
|
PWP1 homolog (S. cerevisiae) |
| chr13_-_33667922 | 0.51 |
ENSDART00000141631
|
si:dkey-76k16.5
|
si:dkey-76k16.5 |
| chr15_-_26931541 | 0.51 |
ENSDART00000027563
|
ccdc9
|
coiled-coil domain containing 9 |
| chr18_-_16885362 | 0.50 |
ENSDART00000132778
|
swap70b
|
switching B cell complex subunit SWAP70b |
| chr25_-_7709797 | 0.50 |
ENSDART00000157076
|
prdm11
|
PR domain containing 11 |
| chr4_-_16853464 | 0.50 |
ENSDART00000125743
ENSDART00000164570 |
slc25a3a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3a |
| chr3_-_26921475 | 0.50 |
ENSDART00000130281
|
ciita
|
class II, major histocompatibility complex, transactivator |
| chr16_-_38629208 | 0.49 |
ENSDART00000126705
|
eif3ea
|
eukaryotic translation initiation factor 3, subunit E, a |
| chr20_-_630637 | 0.49 |
ENSDART00000160454
|
si:dkey-121j17.5
|
si:dkey-121j17.5 |
| chr5_-_68826177 | 0.48 |
ENSDART00000136605
|
si:ch211-283h6.4
|
si:ch211-283h6.4 |
| chr11_+_24820542 | 0.48 |
ENSDART00000135443
|
kdm5ba
|
lysine (K)-specific demethylase 5Ba |
| chr5_+_13245589 | 0.48 |
ENSDART00000193618
|
CR933528.1
|
|
| chr4_+_57101749 | 0.48 |
ENSDART00000135121
|
si:ch211-238e22.4
|
si:ch211-238e22.4 |
| chr6_-_53885514 | 0.47 |
ENSDART00000173812
ENSDART00000127144 |
cacna2d2a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2a |
| chr10_-_32494499 | 0.47 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr6_-_51556308 | 0.46 |
ENSDART00000149033
|
rbl1
|
retinoblastoma-like 1 (p107) |
| chr4_-_69550757 | 0.45 |
ENSDART00000168344
|
znf1101
|
zinc finger protein 1101 |
| chr10_+_40742685 | 0.44 |
ENSDART00000184858
ENSDART00000140300 |
taar19e
|
trace amine associated receptor 19e |
| chr11_-_889845 | 0.43 |
ENSDART00000162152
|
atg7
|
ATG7 autophagy related 7 homolog (S. cerevisiae) |
| chr7_+_72003301 | 0.43 |
ENSDART00000012918
ENSDART00000182268 ENSDART00000185750 |
psmd9
|
proteasome 26S subunit, non-ATPase 9 |
| chr5_+_5398966 | 0.42 |
ENSDART00000139553
|
mapkap1
|
mitogen-activated protein kinase associated protein 1 |
| chr10_+_44146287 | 0.42 |
ENSDART00000162176
|
grk3
|
G protein-coupled receptor kinase 3 |
| chr23_-_4925641 | 0.41 |
ENSDART00000140861
ENSDART00000060718 |
taz
|
tafazzin |
| chr7_-_56559717 | 0.39 |
ENSDART00000161115
|
CR382281.1
|
|
| chr7_-_28568238 | 0.39 |
ENSDART00000173927
|
tmem9b
|
TMEM9 domain family, member B |
| chr11_+_26403334 | 0.39 |
ENSDART00000171781
|
fer1l4
|
fer-1-like family member 4 |
| chr10_-_32494304 | 0.38 |
ENSDART00000028161
|
uvrag
|
UV radiation resistance associated gene |
| chr18_+_3243292 | 0.37 |
ENSDART00000166580
|
pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr22_+_9522971 | 0.37 |
ENSDART00000110048
|
strip1
|
striatin interacting protein 1 |
| chr19_+_15440841 | 0.37 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr4_-_20292821 | 0.36 |
ENSDART00000136069
ENSDART00000192504 |
cacna2d4a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4a |
| chr3_+_32125452 | 0.35 |
ENSDART00000110396
|
zgc:194125
|
zgc:194125 |
| chr8_-_45867358 | 0.35 |
ENSDART00000132810
|
adam9
|
ADAM metallopeptidase domain 9 |
| chr12_-_13966184 | 0.34 |
ENSDART00000066368
|
klhl11
|
kelch-like family member 11 |
Network of associatons between targets according to the STRING database.
First level regulatory network of zbtb16a+zbtb16b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 21.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 3.2 | 9.6 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 2.0 | 7.8 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.8 | 3.2 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.7 | 2.2 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.7 | 2.0 | GO:0090248 | hindbrain structural organization(GO:0021577) dorsal fin morphogenesis(GO:0035142) cell motility involved in somitogenic axis elongation(GO:0090247) cell migration involved in somitogenic axis elongation(GO:0090248) |
| 0.6 | 3.2 | GO:0030328 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 0.6 | 6.0 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.6 | 1.7 | GO:0044038 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.5 | 6.6 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.4 | 1.2 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.4 | 1.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.4 | 1.9 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.3 | 9.7 | GO:0019835 | cytolysis(GO:0019835) |
| 0.3 | 2.2 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.3 | 1.7 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.3 | 1.4 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.3 | 2.3 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) |
| 0.2 | 2.5 | GO:0043545 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.2 | 7.8 | GO:0021955 | central nervous system neuron axonogenesis(GO:0021955) |
| 0.2 | 0.7 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.2 | 0.7 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.2 | 0.9 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.2 | 0.6 | GO:0072068 | proximal straight tubule development(GO:0072020) distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 0.2 | 3.6 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.2 | 1.7 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.2 | 0.8 | GO:2000660 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.2 | 0.9 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.2 | 1.3 | GO:2001032 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.2 | 3.7 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.1 | 0.4 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.1 | 1.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.7 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.1 | 0.7 | GO:0051292 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.1 | 2.7 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 6.2 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 1.2 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 2.7 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.1 | 0.5 | GO:1904867 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 0.1 | 1.5 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 2.7 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 0.4 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.1 | 1.5 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 2.2 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 1.9 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.1 | 3.8 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.1 | 1.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.5 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 3.2 | GO:0001878 | response to yeast(GO:0001878) |
| 0.1 | 0.7 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 2.5 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 0.2 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.1 | 3.7 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 0.8 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 1.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 1.9 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.5 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 0.4 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 1.7 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.1 | 1.6 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.1 | 15.9 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 1.2 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.4 | GO:0003315 | heart rudiment formation(GO:0003315) |
| 0.0 | 1.4 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 0.4 | GO:0045823 | positive regulation of heart contraction(GO:0045823) |
| 0.0 | 2.4 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 3.2 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.5 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.4 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0048855 | adenohypophysis morphogenesis(GO:0048855) |
| 0.0 | 0.5 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 1.6 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.7 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.3 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.5 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 4.4 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.7 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.1 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.0 | 0.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 1.6 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 0.7 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 1.2 | GO:0008203 | cholesterol metabolic process(GO:0008203) secondary alcohol metabolic process(GO:1902652) |
| 0.0 | 1.2 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.8 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.8 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 1.0 | GO:0042113 | B cell activation(GO:0042113) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 21.2 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.8 | 12.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.5 | 2.9 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.4 | 2.5 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.4 | 1.1 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.3 | 1.4 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.3 | 3.8 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.2 | 2.7 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.2 | 0.7 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.2 | 2.3 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.1 | 1.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.4 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 0.5 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.1 | 3.2 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 0.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 2.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 9.6 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.9 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 1.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 1.2 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 1.6 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 3.7 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.1 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 2.0 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 12.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 5.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.2 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.6 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.1 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.3 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.2 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 1.0 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 1.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 3.3 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 8.3 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.6 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 21.2 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 2.0 | 6.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 2.0 | 7.8 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 1.3 | 7.8 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 1.3 | 5.1 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 1.1 | 9.6 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.9 | 2.7 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.8 | 2.5 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.7 | 4.2 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.6 | 3.8 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.6 | 3.2 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.6 | 3.0 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.6 | 2.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.5 | 13.2 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.5 | 4.5 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.4 | 11.1 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.4 | 4.0 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.3 | 3.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.3 | 1.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 0.7 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.2 | 0.9 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.2 | 0.7 | GO:0047453 | ATP-dependent NAD(P)H-hydrate dehydratase activity(GO:0047453) ADP-dependent NAD(P)H-hydrate dehydratase activity(GO:0052855) |
| 0.2 | 1.3 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.2 | 0.8 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.2 | 2.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 1.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.2 | 0.5 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.2 | 1.0 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.2 | 5.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 1.5 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 2.9 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 2.8 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 2.1 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 0.7 | GO:0097363 | protein N-acetylglucosaminyltransferase activity(GO:0016262) protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 1.7 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.8 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 1.9 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 1.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.5 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 1.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 6.2 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 0.7 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.1 | 1.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.4 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 2.2 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 0.4 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 0.6 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 0.3 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 1.0 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.5 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.6 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.1 | GO:0001635 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.0 | 1.7 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 3.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.7 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.9 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.9 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.3 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 1.8 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 3.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 2.2 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 1.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.4 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 1.0 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 3.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 5.3 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.6 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.5 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.1 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 11.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.2 | 0.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 3.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 23.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 0.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 3.7 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 2.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 2.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.4 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.2 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.7 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 2.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 22.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 2.8 | 11.1 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 1.9 | 9.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.5 | 13.1 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.3 | 7.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.2 | 2.8 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 2.9 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 4.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 3.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.5 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 1.0 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 2.7 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 1.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 2.0 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 3.7 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 0.4 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 2.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 0.9 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 0.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.6 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.9 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.4 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 2.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 1.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.4 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |