Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
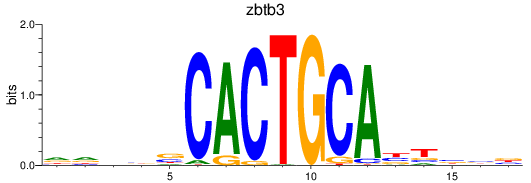
Results for zbtb3
Z-value: 1.06
Transcription factors associated with zbtb3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zbtb3
|
ENSDARG00000036235 | zinc finger and BTB domain containing 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zbtb3 | dr11_v1_chr14_+_35405518_35405518 | -0.25 | 1.5e-02 | Click! |
Activity profile of zbtb3 motif
Sorted Z-values of zbtb3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_52398205 | 123.68 |
ENSDART00000143225
|
si:ch211-217k17.9
|
si:ch211-217k17.9 |
| chr3_-_61185746 | 16.11 |
ENSDART00000028219
|
pvalb4
|
parvalbumin 4 |
| chr16_-_42872571 | 12.68 |
ENSDART00000154757
ENSDART00000102345 |
txnipb
|
thioredoxin interacting protein b |
| chr2_+_68789 | 10.27 |
ENSDART00000058569
|
cldn1
|
claudin 1 |
| chr15_-_2640966 | 9.19 |
ENSDART00000063320
|
cldne
|
claudin e |
| chr11_+_10548171 | 8.39 |
ENSDART00000191497
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr9_+_307863 | 7.53 |
ENSDART00000163474
|
stac3
|
SH3 and cysteine rich domain 3 |
| chr4_-_13502549 | 5.76 |
ENSDART00000140366
|
si:ch211-266a5.12
|
si:ch211-266a5.12 |
| chr20_+_25340814 | 5.74 |
ENSDART00000063028
|
ctgfa
|
connective tissue growth factor a |
| chr4_+_38550788 | 4.53 |
ENSDART00000157412
|
si:ch211-209n20.1
|
si:ch211-209n20.1 |
| chr13_-_2215213 | 4.42 |
ENSDART00000129773
|
mlip
|
muscular LMNA-interacting protein |
| chr19_+_7878122 | 4.39 |
ENSDART00000131917
|
si:dkeyp-85e10.2
|
si:dkeyp-85e10.2 |
| chr24_+_6107901 | 4.35 |
ENSDART00000156419
|
si:ch211-37e10.2
|
si:ch211-37e10.2 |
| chr2_-_985417 | 4.32 |
ENSDART00000140540
|
si:ch211-241e1.3
|
si:ch211-241e1.3 |
| chr19_+_917852 | 4.21 |
ENSDART00000082466
|
tgfbr2a
|
transforming growth factor beta receptor 2a |
| chr7_-_35432901 | 4.16 |
ENSDART00000026712
|
mmp2
|
matrix metallopeptidase 2 |
| chr12_+_48220584 | 4.04 |
ENSDART00000164392
|
lrrc20
|
leucine rich repeat containing 20 |
| chr6_+_7507390 | 3.96 |
ENSDART00000149285
|
erbb3a
|
erb-b2 receptor tyrosine kinase 3a |
| chr17_-_868004 | 3.76 |
ENSDART00000112803
|
wdr20a
|
WD repeat domain 20a |
| chr25_-_33275666 | 3.62 |
ENSDART00000193605
|
CABZ01046954.1
|
|
| chr7_+_6879534 | 3.51 |
ENSDART00000157731
|
zgc:175248
|
zgc:175248 |
| chr25_+_25453221 | 2.98 |
ENSDART00000176822
|
rassf7a
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7a |
| chr19_-_48010490 | 2.92 |
ENSDART00000159938
|
FBXL19
|
zgc:158376 |
| chr1_+_58079053 | 2.90 |
ENSDART00000141678
|
si:ch211-114l13.3
|
si:ch211-114l13.3 |
| chr4_+_12966640 | 2.84 |
ENSDART00000113357
|
vhll
|
von Hippel-Lindau tumor suppressor like |
| chr21_-_34951265 | 2.84 |
ENSDART00000135222
|
lipia
|
lipase, member Ia |
| chr16_-_32013913 | 2.77 |
ENSDART00000030282
ENSDART00000138701 |
gstk1
|
glutathione S-transferase kappa 1 |
| chr3_+_49397115 | 2.70 |
ENSDART00000176042
|
tecra
|
trans-2,3-enoyl-CoA reductase a |
| chr6_-_39006449 | 2.63 |
ENSDART00000150885
|
vdrb
|
vitamin D receptor b |
| chr18_-_50839033 | 2.60 |
ENSDART00000169773
|
ddb1
|
damage-specific DNA binding protein 1 |
| chr14_-_1355544 | 2.55 |
ENSDART00000060417
|
cetn4
|
centrin 4 |
| chr3_-_34069637 | 2.47 |
ENSDART00000151588
|
ighv9-1
|
immunoglobulin heavy variable 9-1 |
| chr3_-_3019979 | 2.44 |
ENSDART00000111174
|
zgc:172090
|
zgc:172090 |
| chr2_+_25198648 | 2.40 |
ENSDART00000110922
|
ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr17_+_52524452 | 2.38 |
ENSDART00000016614
|
dlst
|
dihydrolipoamide S-succinyltransferase |
| chr4_-_68913650 | 2.29 |
ENSDART00000184297
|
si:dkey-264f17.5
|
si:dkey-264f17.5 |
| chr8_+_54135642 | 2.20 |
ENSDART00000170712
|
brpf1
|
bromodomain and PHD finger containing, 1 |
| chr3_-_5353442 | 2.17 |
ENSDART00000188808
|
trim35-7
|
tripartite motif containing 35-7 |
| chr11_-_42230491 | 2.00 |
ENSDART00000164423
|
CABZ01030862.1
|
|
| chr23_-_7674902 | 1.93 |
ENSDART00000185612
ENSDART00000180524 |
plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr1_-_54425791 | 1.93 |
ENSDART00000039911
|
pkd1a
|
polycystic kidney disease 1a |
| chr24_-_26383978 | 1.90 |
ENSDART00000031426
|
skilb
|
SKI-like proto-oncogene b |
| chr9_+_48088031 | 1.77 |
ENSDART00000073870
|
si:ch73-54b5.2
|
si:ch73-54b5.2 |
| chr15_-_19705707 | 1.77 |
ENSDART00000047643
|
sytl2b
|
synaptotagmin-like 2b |
| chr2_+_47718605 | 1.66 |
ENSDART00000189180
ENSDART00000148824 |
mbnl1
|
muscleblind-like splicing regulator 1 |
| chr1_+_59007536 | 1.66 |
ENSDART00000165339
|
CDC37
|
cell division cycle 37 |
| chr1_-_633356 | 1.54 |
ENSDART00000171019
|
appa
|
amyloid beta (A4) precursor protein a |
| chr8_-_1884955 | 1.47 |
ENSDART00000081563
|
si:dkey-178e17.3
|
si:dkey-178e17.3 |
| chr7_-_26270014 | 1.47 |
ENSDART00000079347
|
serpine1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr3_+_61185660 | 1.30 |
ENSDART00000167114
|
CR352263.1
|
|
| chr6_+_12527725 | 1.26 |
ENSDART00000149328
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr10_+_31951338 | 1.24 |
ENSDART00000019416
|
lhfpl6
|
LHFPL tetraspan subfamily member 6 |
| chr8_+_22404981 | 1.17 |
ENSDART00000185211
ENSDART00000099972 |
si:dkey-23c22.7
si:dkey-23c22.9
|
si:dkey-23c22.7 si:dkey-23c22.9 |
| chr18_+_37015185 | 1.16 |
ENSDART00000191305
|
sipa1l3
|
signal-induced proliferation-associated 1 like 3 |
| chr19_-_35439237 | 1.15 |
ENSDART00000145883
|
anln
|
anillin, actin binding protein |
| chr1_+_532766 | 1.08 |
ENSDART00000179731
ENSDART00000060944 |
mrpl39
|
mitochondrial ribosomal protein L39 |
| chr16_+_10422836 | 1.06 |
ENSDART00000161568
|
ino80e
|
INO80 complex subunit E |
| chr19_-_5264517 | 1.05 |
ENSDART00000001821
|
prf1.3
|
perforin 1.3 |
| chr8_-_8446668 | 1.04 |
ENSDART00000132700
|
cdk16
|
cyclin-dependent kinase 16 |
| chr8_+_26293673 | 1.02 |
ENSDART00000144977
|
mgll
|
monoglyceride lipase |
| chr21_+_35296246 | 1.01 |
ENSDART00000076750
|
il12bb
|
interleukin 12B, b |
| chr7_+_30051880 | 1.00 |
ENSDART00000075609
|
pstpip1b
|
proline-serine-threonine phosphatase interacting protein 1b |
| chr1_+_9153141 | 1.00 |
ENSDART00000081343
|
plk1
|
polo-like kinase 1 (Drosophila) |
| chr20_+_27713210 | 0.99 |
ENSDART00000132222
|
zbtb1
|
zinc finger and BTB domain containing 1 |
| chr3_+_53240562 | 0.87 |
ENSDART00000031234
|
stxbp2
|
syntaxin binding protein 2 |
| chr7_+_24522308 | 0.85 |
ENSDART00000173542
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr2_-_27329214 | 0.72 |
ENSDART00000145835
|
tmx3a
|
thioredoxin related transmembrane protein 3a |
| chr8_+_30921403 | 0.70 |
ENSDART00000043251
|
derl3
|
derlin 3 |
| chr7_+_24523017 | 0.64 |
ENSDART00000077047
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr12_+_48683208 | 0.61 |
ENSDART00000188582
|
mmp21
|
matrix metallopeptidase 21 |
| chr18_+_33521609 | 0.60 |
ENSDART00000147592
|
v2rh7
|
vomeronasal 2 receptor, h7 |
| chr7_-_25895189 | 0.60 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr13_+_30035253 | 0.59 |
ENSDART00000181303
ENSDART00000057525 ENSDART00000136622 |
dnajb12a
|
DnaJ (Hsp40) homolog, subfamily B, member 12a |
| chr19_-_19442983 | 0.55 |
ENSDART00000052649
|
sb:cb649
|
sb:cb649 |
| chr16_-_30969294 | 0.53 |
ENSDART00000147890
|
dgat1b
|
diacylglycerol O-acyltransferase 1b |
| chr11_+_1805421 | 0.53 |
ENSDART00000173143
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr8_-_53488832 | 0.47 |
ENSDART00000191801
|
chdh
|
choline dehydrogenase |
| chr14_+_24845941 | 0.46 |
ENSDART00000187513
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr24_+_21720304 | 0.42 |
ENSDART00000147250
ENSDART00000048273 |
pan3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr17_+_34215886 | 0.37 |
ENSDART00000186775
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr3_+_8826905 | 0.35 |
ENSDART00000167474
ENSDART00000168088 |
si:dkeyp-30d6.2
|
si:dkeyp-30d6.2 |
| chr9_-_27738110 | 0.30 |
ENSDART00000060347
|
crygs2
|
crystallin, gamma S2 |
| chr7_-_34225011 | 0.27 |
ENSDART00000049588
|
crybgx
|
crystallin beta gamma X |
| chr9_-_28274932 | 0.27 |
ENSDART00000137582
ENSDART00000146932 |
creb1b
|
cAMP responsive element binding protein 1b |
| chr14_+_24934736 | 0.27 |
ENSDART00000191821
|
ppargc1b
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr1_-_22687913 | 0.25 |
ENSDART00000168171
|
fgfbp2b
|
fibroblast growth factor binding protein 2b |
| chr6_-_51556308 | 0.22 |
ENSDART00000149033
|
rbl1
|
retinoblastoma-like 1 (p107) |
| chr20_+_39375280 | 0.18 |
ENSDART00000172434
|
pimr131
|
Pim proto-oncogene, serine/threonine kinase, related 131 |
| chr17_-_29311835 | 0.17 |
ENSDART00000104224
|
tecpr2
|
tectonin beta-propeller repeat containing 2 |
| chr24_+_12740848 | 0.14 |
ENSDART00000060825
|
si:ch211-196f5.2
|
si:ch211-196f5.2 |
| chr22_-_5051689 | 0.13 |
ENSDART00000113521
|
matk
|
megakaryocyte-associated tyrosine kinase |
| chr7_+_39079071 | 0.10 |
ENSDART00000181845
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr23_-_29878643 | 0.10 |
ENSDART00000058407
|
slc25a33
|
solute carrier family 25 (pyrimidine nucleotide carrier), member 33 |
| chr16_-_39131666 | 0.09 |
ENSDART00000075517
|
gdf6a
|
growth differentiation factor 6a |
| chr22_-_18387059 | 0.08 |
ENSDART00000007769
|
tssk6
|
testis-specific serine kinase 6 |
| chr17_+_30450163 | 0.07 |
ENSDART00000104257
|
lpin1
|
lipin 1 |
| chr5_+_9297585 | 0.04 |
ENSDART00000170557
|
si:ch73-194c23.1
|
si:ch73-194c23.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of zbtb3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.5 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.8 | 2.4 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.5 | 1.5 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.5 | 1.9 | GO:0044246 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) |
| 0.4 | 2.5 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.3 | 16.8 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.3 | 1.2 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.2 | 4.2 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.2 | 0.7 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.2 | 2.8 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.2 | 2.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.2 | 0.5 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 1.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 1.0 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 2.7 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 1.9 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 0.8 | GO:2000406 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.1 | 4.2 | GO:0071559 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.1 | 4.3 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 2.6 | GO:0003146 | heart jogging(GO:0003146) |
| 0.0 | 2.2 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 1.1 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 2.4 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.1 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.6 | GO:0051788 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.0 | 7.6 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.1 | GO:0060220 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.0 | 1.0 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.9 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.6 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.6 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.7 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 2.7 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 1.5 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.4 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.2 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 7.6 | GO:0016567 | protein ubiquitination(GO:0016567) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.8 | GO:0030891 | VCB complex(GO:0030891) |
| 0.4 | 5.8 | GO:0030315 | T-tubule(GO:0030315) |
| 0.3 | 1.0 | GO:0070743 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.2 | 2.4 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.2 | 4.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 1.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.4 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 0.7 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 16.8 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 2.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 4.4 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 2.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 2.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.1 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 4.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 2.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 4.2 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 2.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 2.8 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 4.2 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 0.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 8.4 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.0 | GO:0000922 | spindle pole(GO:0000922) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.4 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 1.1 | 4.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.8 | 2.4 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.8 | 4.0 | GO:0038131 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.7 | 4.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.5 | 2.6 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.3 | 2.8 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.3 | 1.0 | GO:0019972 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.2 | 0.7 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.2 | 12.7 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.2 | 2.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 0.7 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.1 | 2.8 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 1.0 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.5 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 1.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 2.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 1.9 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 3.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 16.6 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 2.4 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 18.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.9 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 1.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 10.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.3 | 4.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 1.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.0 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 10.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.6 | 4.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 2.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 2.6 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 1.0 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.7 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.7 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |