Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
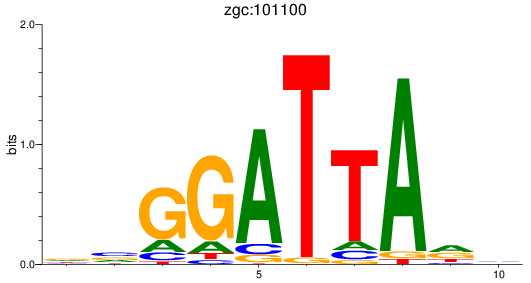
Results for zgc:101100
Z-value: 0.57
Transcription factors associated with zgc:101100
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zgc
|
ENSDARG00000021959 | 101100 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zgc:101100 | dr11_v1_chr8_+_20488322_20488322 | 0.63 | 1.0e-11 | Click! |
Activity profile of zgc:101100 motif
Sorted Z-values of zgc:101100 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_3326885 | 5.80 |
ENSDART00000104866
|
ldhbb
|
lactate dehydrogenase Bb |
| chr3_-_49566364 | 5.62 |
ENSDART00000161507
|
zgc:153426
|
zgc:153426 |
| chr2_-_30784198 | 5.22 |
ENSDART00000182523
ENSDART00000147355 |
rgs20
|
regulator of G protein signaling 20 |
| chr20_-_29475172 | 4.92 |
ENSDART00000183164
|
scg5
|
secretogranin V |
| chr12_+_41991635 | 4.92 |
ENSDART00000186161
ENSDART00000192510 |
TCERG1L
|
transcription elongation regulator 1 like |
| chr24_-_38079261 | 4.72 |
ENSDART00000105662
|
crp1
|
C-reactive protein 1 |
| chr22_+_5103349 | 4.63 |
ENSDART00000083474
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr20_-_18736281 | 4.48 |
ENSDART00000142837
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr20_+_53441935 | 4.01 |
ENSDART00000175214
|
apobb.2
|
apolipoprotein Bb, tandem duplicate 2 |
| chr16_+_32059785 | 3.93 |
ENSDART00000134459
|
si:dkey-40m6.8
|
si:dkey-40m6.8 |
| chr5_-_50748081 | 3.80 |
ENSDART00000113985
|
mctp1a
|
multiple C2 domains, transmembrane 1a |
| chr7_-_49594995 | 3.79 |
ENSDART00000174161
ENSDART00000109147 |
brsk2b
|
BR serine/threonine kinase 2b |
| chr7_-_57933736 | 3.69 |
ENSDART00000142580
|
ank2b
|
ankyrin 2b, neuronal |
| chr19_+_1097393 | 3.65 |
ENSDART00000168218
|
CABZ01111953.1
|
|
| chr12_-_10705916 | 3.41 |
ENSDART00000164038
|
FO704786.1
|
|
| chr22_+_34701848 | 3.39 |
ENSDART00000082066
|
atpv0e2
|
ATPase H+ transporting V0 subunit e2 |
| chr1_-_45920632 | 3.38 |
ENSDART00000140890
|
pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr16_-_31675669 | 3.36 |
ENSDART00000168848
ENSDART00000158331 |
c1r
|
complement component 1, r subcomponent |
| chr9_+_13714379 | 3.16 |
ENSDART00000017593
ENSDART00000145503 |
tmem237a
|
transmembrane protein 237a |
| chr10_+_6013076 | 3.10 |
ENSDART00000167613
ENSDART00000159216 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr3_+_34220194 | 3.08 |
ENSDART00000145859
|
slc25a23b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23b |
| chr15_-_23793641 | 2.91 |
ENSDART00000122891
|
tmem97
|
transmembrane protein 97 |
| chr3_+_34919810 | 2.85 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr5_+_52067723 | 2.73 |
ENSDART00000166902
|
setbp1
|
SET binding protein 1 |
| chr10_+_17026870 | 2.30 |
ENSDART00000184529
ENSDART00000157480 |
CR855996.2
|
|
| chr5_+_66355153 | 2.18 |
ENSDART00000082745
|
si:ch211-261c8.5
|
si:ch211-261c8.5 |
| chr1_+_55758257 | 2.12 |
ENSDART00000139312
|
tecrb
|
trans-2,3-enoyl-CoA reductase b |
| chr1_+_47585700 | 1.87 |
ENSDART00000153746
ENSDART00000084457 |
sh3pxd2aa
|
SH3 and PX domains 2Aa |
| chr15_-_26887028 | 1.75 |
ENSDART00000156292
|
si:dkey-243i1.1
|
si:dkey-243i1.1 |
| chr23_-_35694461 | 1.61 |
ENSDART00000185884
|
tuba1c
|
tubulin, alpha 1c |
| chr2_+_13050225 | 1.59 |
ENSDART00000182237
|
BX324233.1
|
|
| chr23_-_10137322 | 1.55 |
ENSDART00000142442
|
plxnb1a
|
plexin b1a |
| chr1_+_41690402 | 1.51 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr19_-_40906161 | 1.43 |
ENSDART00000004514
|
bet1
|
Bet1 golgi vesicular membrane trafficking protein |
| chr11_+_30296332 | 1.41 |
ENSDART00000192843
|
ugt1b7
|
UDP glucuronosyltransferase 1 family, polypeptide B7 |
| chr23_-_21797517 | 1.38 |
ENSDART00000110041
|
lrrc38a
|
leucine rich repeat containing 38a |
| chr7_+_20031202 | 1.32 |
ENSDART00000052904
|
slc16a13
|
solute carrier family 16, member 13 (monocarboxylic acid transporter 13) |
| chr10_+_2841205 | 1.29 |
ENSDART00000131505
ENSDART00000055869 |
ykt6
|
YKT6 v-SNARE homolog (S. cerevisiae) |
| chr11_-_3366782 | 1.28 |
ENSDART00000127982
|
suox
|
sulfite oxidase |
| chr19_-_32518556 | 1.23 |
ENSDART00000103410
|
zbtb8b
|
zinc finger and BTB domain containing 8B |
| chr6_-_17999776 | 1.21 |
ENSDART00000183048
ENSDART00000181577 ENSDART00000170597 |
rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr17_-_15149192 | 1.19 |
ENSDART00000180511
ENSDART00000103405 |
gch1
|
GTP cyclohydrolase 1 |
| chr5_-_28625515 | 1.12 |
ENSDART00000190782
ENSDART00000179736 ENSDART00000131729 |
tnc
|
tenascin C |
| chr5_+_31791001 | 1.12 |
ENSDART00000043010
|
slc25a25b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25b |
| chr10_-_7988396 | 1.07 |
ENSDART00000141445
ENSDART00000024282 |
ewsr1a
|
EWS RNA-binding protein 1a |
| chr18_+_19975787 | 1.00 |
ENSDART00000138103
|
skor1b
|
SKI family transcriptional corepressor 1b |
| chr6_-_39653972 | 1.00 |
ENSDART00000155859
|
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr17_+_35097024 | 0.99 |
ENSDART00000026152
|
asap2a
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2a |
| chr17_-_50010121 | 0.98 |
ENSDART00000122747
|
tmem30aa
|
transmembrane protein 30Aa |
| chr23_-_20345473 | 0.97 |
ENSDART00000140935
|
si:rp71-17i16.6
|
si:rp71-17i16.6 |
| chr19_+_24068223 | 0.95 |
ENSDART00000141351
ENSDART00000100420 |
pex11b
|
peroxisomal biogenesis factor 11 beta |
| chr24_-_25004553 | 0.90 |
ENSDART00000080997
ENSDART00000136860 |
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr2_-_48171441 | 0.84 |
ENSDART00000123040
|
pfkpb
|
phosphofructokinase, platelet b |
| chr13_-_50614639 | 0.82 |
ENSDART00000170527
|
vent
|
ventral expressed homeobox |
| chr18_+_7639401 | 0.79 |
ENSDART00000092416
|
rabl2
|
RAB, member of RAS oncogene family-like 2 |
| chr11_+_25583950 | 0.79 |
ENSDART00000111961
|
ccdc120
|
coiled-coil domain containing 120 |
| chr14_-_2933185 | 0.78 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr7_-_12821277 | 0.78 |
ENSDART00000091584
|
zgc:158785
|
zgc:158785 |
| chr21_+_44773237 | 0.77 |
ENSDART00000084780
|
rnf121
|
ring finger protein 121 |
| chr13_-_36525982 | 0.76 |
ENSDART00000114744
|
pacs2
|
phosphofurin acidic cluster sorting protein 2 |
| chr15_-_15469079 | 0.74 |
ENSDART00000132637
ENSDART00000004220 |
rab34a
|
RAB34, member RAS oncogene family a |
| chr10_+_28428222 | 0.70 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr8_+_23142946 | 0.69 |
ENSDART00000152933
|
si:ch211-196c10.13
|
si:ch211-196c10.13 |
| chr10_-_26274094 | 0.66 |
ENSDART00000108798
|
dchs1b
|
dachsous cadherin-related 1b |
| chr14_-_46395408 | 0.65 |
ENSDART00000147537
|
bbs7
|
Bardet-Biedl syndrome 7 |
| chr2_+_30786773 | 0.61 |
ENSDART00000019029
ENSDART00000145681 |
atp6v1h
|
ATPase H+ transporting V1 subunit H |
| chr16_-_17188294 | 0.57 |
ENSDART00000165883
|
opn9
|
opsin 9 |
| chr1_+_50613868 | 0.56 |
ENSDART00000111114
|
HERC5
|
si:ch73-190m4.1 |
| chr9_-_12682848 | 0.53 |
ENSDART00000138360
|
myo10l3
|
myosin X-like 3 |
| chr6_-_2222707 | 0.51 |
ENSDART00000022179
|
prkag1
|
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr7_-_17780048 | 0.50 |
ENSDART00000183336
|
si:dkey-106g10.7
|
si:dkey-106g10.7 |
| chr7_-_17779644 | 0.50 |
ENSDART00000128504
|
si:dkey-106g10.7
|
si:dkey-106g10.7 |
| chr9_-_29544720 | 0.42 |
ENSDART00000130317
|
arhgap20
|
Rho GTPase activating protein 20 |
| chr21_-_15046065 | 0.42 |
ENSDART00000178507
|
mmp17a
|
matrix metallopeptidase 17a |
| chr22_+_438714 | 0.41 |
ENSDART00000136491
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr21_-_41369370 | 0.40 |
ENSDART00000159290
|
cpeb4b
|
cytoplasmic polyadenylation element binding protein 4b |
| chr5_+_24287927 | 0.40 |
ENSDART00000143563
|
zdhhc23a
|
zinc finger, DHHC-type containing 23a |
| chr2_+_30787128 | 0.38 |
ENSDART00000189233
|
atp6v1h
|
ATPase H+ transporting V1 subunit H |
| chr15_+_5973909 | 0.38 |
ENSDART00000126886
ENSDART00000189618 |
igsf5b
|
immunoglobulin superfamily, member 5b |
| chr2_-_9696283 | 0.34 |
ENSDART00000165712
|
selenot1a
|
selenoprotein T, 1a |
| chr14_+_32837914 | 0.34 |
ENSDART00000158888
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr23_+_6544453 | 0.34 |
ENSDART00000005373
|
spo11
|
SPO11 meiotic protein covalently bound to DSB |
| chr6_-_33924883 | 0.33 |
ENSDART00000132762
ENSDART00000148142 ENSDART00000142213 |
akr1a1b
|
aldo-keto reductase family 1, member A1b (aldehyde reductase) |
| chr20_+_34671386 | 0.31 |
ENSDART00000152836
ENSDART00000138226 |
elp3
|
elongator acetyltransferase complex subunit 3 |
| chr4_+_65271779 | 0.31 |
ENSDART00000134257
|
znf1027
|
zinc finger protein 1027 |
| chr3_-_38918697 | 0.30 |
ENSDART00000145630
|
si:dkey-106c17.2
|
si:dkey-106c17.2 |
| chr11_+_15613218 | 0.29 |
ENSDART00000066033
|
gdf11
|
growth differentiation factor 11 |
| chr3_-_8510201 | 0.29 |
ENSDART00000009151
|
CABZ01064671.1
|
|
| chr13_-_33153889 | 0.28 |
ENSDART00000141451
|
TC2N
|
tandem C2 domains, nuclear |
| chr22_+_35275468 | 0.27 |
ENSDART00000189516
ENSDART00000181572 ENSDART00000165353 ENSDART00000185352 |
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr8_+_2656231 | 0.26 |
ENSDART00000160833
|
fam102aa
|
family with sequence similarity 102, member Aa |
| chr16_+_16265850 | 0.24 |
ENSDART00000181265
|
setd2
|
SET domain containing 2 |
| chr16_+_50163352 | 0.21 |
ENSDART00000153890
ENSDART00000060522 |
si:ch211-231m23.4
|
si:ch211-231m23.4 |
| chr18_-_25646286 | 0.19 |
ENSDART00000099511
ENSDART00000186890 |
si:ch211-13k12.2
|
si:ch211-13k12.2 |
| chr6_-_59381391 | 0.18 |
ENSDART00000157066
|
ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr25_-_17503393 | 0.17 |
ENSDART00000061726
|
trim35-40
|
tripartite motif containing 35-40 |
| chr6_+_41186320 | 0.16 |
ENSDART00000025241
|
opn1mw2
|
opsin 1 (cone pigments), medium-wave-sensitive, 2 |
| chr22_-_14696667 | 0.13 |
ENSDART00000180379
|
lrp1ba
|
low density lipoprotein receptor-related protein 1Ba |
| chr25_-_32381642 | 0.11 |
ENSDART00000133872
ENSDART00000006124 |
cops2
|
COP9 signalosome subunit 2 |
| chr14_-_22152064 | 0.11 |
ENSDART00000115422
|
nudcd2
|
NudC domain containing 2 |
| chr2_+_30531726 | 0.10 |
ENSDART00000146518
|
ankrd33bb
|
ankyrin repeat domain 33Bb |
| chr5_-_38107741 | 0.05 |
ENSDART00000156853
|
si:ch211-284e13.14
|
si:ch211-284e13.14 |
Network of associatons between targets according to the STRING database.
First level regulatory network of zgc:101100
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0010142 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.8 | 4.0 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.7 | 3.7 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.4 | 4.6 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.4 | 1.1 | GO:0014814 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.3 | 1.2 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.3 | 1.1 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.3 | 0.8 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.2 | 3.4 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.2 | 0.7 | GO:1903673 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.2 | 1.0 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.2 | 1.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 1.0 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.1 | 4.3 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.3 | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridine biosynthesis.(GO:0002926) |
| 0.1 | 4.9 | GO:0046883 | regulation of hormone secretion(GO:0046883) |
| 0.1 | 0.3 | GO:0090219 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) negative regulation of phospholipid metabolic process(GO:1903726) |
| 0.1 | 0.7 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 1.6 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 3.8 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 0.8 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.1 | 1.9 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.1 | 0.7 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.1 | 2.1 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 3.4 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.1 | 0.6 | GO:0061621 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 1.7 | GO:0007099 | centriole replication(GO:0007099) |
| 0.1 | 2.8 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.3 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.0 | 1.2 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 3.4 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.2 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 1.3 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.4 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.4 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 1.0 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 1.1 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 3.2 | GO:0060027 | convergent extension involved in gastrulation(GO:0060027) |
| 0.0 | 1.0 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.2 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 1.3 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.4 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.0 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.2 | 3.7 | GO:0030315 | T-tubule(GO:0030315) |
| 0.2 | 3.4 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.2 | 1.4 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.2 | 1.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 1.0 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 1.9 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.0 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 0.7 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 1.6 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 4.9 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 2.2 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 0.6 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 3.8 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.4 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 1.0 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 3.8 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 3.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.8 | 5.8 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.4 | 4.0 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.4 | 4.6 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.2 | 1.9 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 3.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.2 | 3.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 3.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 2.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 1.3 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.1 | 4.2 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.1 | 0.3 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.6 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 1.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.0 | 0.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 2.1 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.0 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.3 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.5 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 2.1 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 1.3 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.4 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 1.3 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 1.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.2 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 1.0 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 3.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.2 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 1.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 3.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.7 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 1.7 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 3.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 5.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 1.0 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.1 | 1.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |