Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
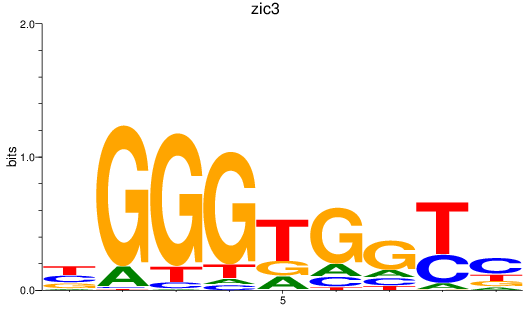
Results for zic3
Z-value: 0.20
Transcription factors associated with zic3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zic3
|
ENSDARG00000071497 | zic family member 3 heterotaxy 1 (odd-paired homolog, Drosophila) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zic3 | dr11_v1_chr14_-_32016615_32016615 | 0.16 | 1.3e-01 | Click! |
Activity profile of zic3 motif
Sorted Z-values of zic3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_27194833 | 3.37 |
ENSDART00000150072
|
si:dkey-85n7.8
|
si:dkey-85n7.8 |
| chr19_+_23982466 | 1.07 |
ENSDART00000080673
|
syt11a
|
synaptotagmin XIa |
| chr22_-_10487490 | 0.88 |
ENSDART00000064798
|
aspn
|
asporin (LRR class 1) |
| chr22_-_37565348 | 0.75 |
ENSDART00000149482
ENSDART00000104478 |
fxr1
|
fragile X mental retardation, autosomal homolog 1 |
| chr12_-_26582607 | 0.51 |
ENSDART00000045186
|
slc16a5a
|
solute carrier family 16 (monocarboxylate transporter), member 5a |
| chr9_+_23770666 | 0.51 |
ENSDART00000182493
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr2_-_30784198 | 0.43 |
ENSDART00000182523
ENSDART00000147355 |
rgs20
|
regulator of G protein signaling 20 |
| chr6_+_27090800 | 0.39 |
ENSDART00000121558
|
atg4b
|
autophagy related 4B, cysteine peptidase |
| chr25_-_34740627 | 0.39 |
ENSDART00000137665
|
frs2b
|
fibroblast growth factor receptor substrate 2b |
| chr5_-_22615087 | 0.39 |
ENSDART00000146035
|
zgc:113208
|
zgc:113208 |
| chr21_-_30415524 | 0.36 |
ENSDART00000101036
|
grpel2
|
GrpE-like 2, mitochondrial |
| chr11_+_34921492 | 0.26 |
ENSDART00000128070
|
gnai2a
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2a |
| chr4_-_56157199 | 0.23 |
ENSDART00000169806
|
si:ch211-207e19.15
|
si:ch211-207e19.15 |
| chr8_+_49096486 | 0.18 |
ENSDART00000192998
|
zgc:56525
|
zgc:56525 |
| chr8_-_40075983 | 0.18 |
ENSDART00000141455
|
ggt1a
|
gamma-glutamyltransferase 1a |
| chr6_+_83873 | 0.15 |
ENSDART00000162145
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr12_+_27117609 | 0.06 |
ENSDART00000076154
|
hoxb8b
|
homeobox B8b |
| chr19_+_9050852 | 0.06 |
ENSDART00000151031
|
ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr21_-_45073764 | 0.04 |
ENSDART00000181390
ENSDART00000063714 |
rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr23_-_14057191 | 0.03 |
ENSDART00000154908
|
adcy6a
|
adenylate cyclase 6a |
| chr20_-_40725142 | 0.03 |
ENSDART00000181279
|
cx32.2
|
connexin 32.2 |
| chr2_+_2918759 | 0.02 |
ENSDART00000161255
|
CABZ01112864.1
|
|
| chr22_-_6988102 | 0.00 |
ENSDART00000185618
|
fgfr1bl
|
fibroblast growth factor receptor 1b, like |
| chr21_-_21465111 | 0.00 |
ENSDART00000141487
|
nectin3b
|
nectin cell adhesion molecule 3b |
Network of associatons between targets according to the STRING database.
First level regulatory network of zic3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.4 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.8 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.3 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 1.1 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.2 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.2 | 3.4 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 0.4 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.8 | GO:0045182 | translation regulator activity(GO:0045182) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |