Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
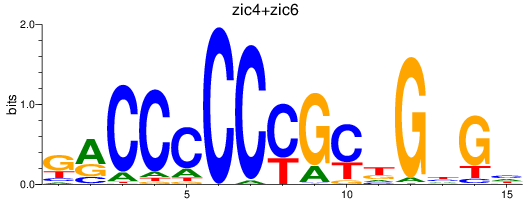
Results for zic4+zic6
Z-value: 0.50
Transcription factors associated with zic4+zic6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zic4
|
ENSDARG00000031307 | zic family member 4 |
|
zic6
|
ENSDARG00000071496 | zic family member 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zic6 | dr11_v1_chr14_+_32022272_32022272 | 0.66 | 6.0e-13 | Click! |
| zic4 | dr11_v1_chr24_+_4978055_4978055 | 0.63 | 1.0e-11 | Click! |
Activity profile of zic4+zic6 motif
Sorted Z-values of zic4+zic6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_17067364 | 5.73 |
ENSDART00000132687
|
rab3c
|
RAB3C, member RAS oncogene family |
| chr8_-_4618653 | 5.49 |
ENSDART00000025535
|
sept5a
|
septin 5a |
| chr16_-_37964325 | 3.92 |
ENSDART00000148801
|
mrap2a
|
melanocortin 2 receptor accessory protein 2a |
| chr5_-_10768258 | 3.80 |
ENSDART00000157043
|
rtn4r
|
reticulon 4 receptor |
| chr19_-_20113696 | 3.64 |
ENSDART00000188813
|
npy
|
neuropeptide Y |
| chr3_+_46724528 | 3.62 |
ENSDART00000181358
|
pde4a
|
phosphodiesterase 4A, cAMP-specific |
| chr13_+_16521898 | 3.45 |
ENSDART00000122557
|
kcnma1a
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1a |
| chr5_+_36611128 | 3.32 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr7_+_30787903 | 3.12 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr16_-_55028740 | 3.12 |
ENSDART00000156368
ENSDART00000161704 |
zgc:114181
|
zgc:114181 |
| chr22_+_4707663 | 3.00 |
ENSDART00000042194
|
cers4a
|
ceramide synthase 4a |
| chr24_-_21172122 | 2.90 |
ENSDART00000154259
|
atp6v1ab
|
ATPase H+ transporting V1 subunit Ab |
| chr8_+_26859639 | 2.81 |
ENSDART00000133440
|
prdm2a
|
PR domain containing 2, with ZNF domain a |
| chr23_+_44741500 | 2.79 |
ENSDART00000166421
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr12_-_41619257 | 2.77 |
ENSDART00000162967
|
dpysl4
|
dihydropyrimidinase-like 4 |
| chr8_-_33114202 | 2.67 |
ENSDART00000098840
|
ralgps1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr14_-_32503363 | 2.38 |
ENSDART00000034883
|
mcf2a
|
MCF.2 cell line derived transforming sequence a |
| chr21_+_3093419 | 2.31 |
ENSDART00000162520
|
SHC3
|
SHC adaptor protein 3 |
| chr10_+_21786656 | 2.28 |
ENSDART00000185851
ENSDART00000167219 |
pcdh1g26
|
protocadherin 1 gamma 26 |
| chr13_+_12739283 | 2.26 |
ENSDART00000102279
|
lingo2b
|
leucine rich repeat and Ig domain containing 2b |
| chr3_-_19091024 | 2.24 |
ENSDART00000188485
ENSDART00000110554 |
grin2ca
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Ca |
| chr20_-_28768109 | 2.24 |
ENSDART00000114611
ENSDART00000182443 |
sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr3_-_25814097 | 2.05 |
ENSDART00000169706
|
ntn1b
|
netrin 1b |
| chr11_-_21586157 | 1.98 |
ENSDART00000190095
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr17_-_4245311 | 1.75 |
ENSDART00000055379
|
gdf3
|
growth differentiation factor 3 |
| chr5_+_20693724 | 1.75 |
ENSDART00000141368
|
si:ch211-240b21.2
|
si:ch211-240b21.2 |
| chr7_-_67842997 | 1.65 |
ENSDART00000169763
|
pmfbp1
|
polyamine modulated factor 1 binding protein 1 |
| chr15_-_26844591 | 1.61 |
ENSDART00000077582
|
pitpnm3
|
PITPNM family member 3 |
| chr13_-_36534380 | 1.46 |
ENSDART00000177857
ENSDART00000181046 ENSDART00000187243 |
srsf5a
|
serine/arginine-rich splicing factor 5a |
| chr6_+_10450000 | 1.42 |
ENSDART00000151288
ENSDART00000187431 ENSDART00000192474 ENSDART00000188214 ENSDART00000184766 ENSDART00000190082 |
kcnh7
|
potassium channel, voltage gated eag related subfamily H, member 7 |
| chr21_-_21465111 | 1.38 |
ENSDART00000141487
|
nectin3b
|
nectin cell adhesion molecule 3b |
| chr25_-_1079417 | 1.37 |
ENSDART00000163134
|
FO907089.1
|
|
| chr1_-_58601636 | 1.34 |
ENSDART00000141143
|
si:ch73-236c18.8
|
si:ch73-236c18.8 |
| chr19_-_17996336 | 1.30 |
ENSDART00000186143
ENSDART00000080751 |
ints8
|
integrator complex subunit 8 |
| chr3_-_19899914 | 1.30 |
ENSDART00000134969
|
rnd2
|
Rho family GTPase 2 |
| chr5_+_31944270 | 1.29 |
ENSDART00000147850
|
ungb
|
uracil DNA glycosylase b |
| chr8_+_40210398 | 1.26 |
ENSDART00000167612
|
rnf34a
|
ring finger protein 34a |
| chr21_-_14664445 | 1.24 |
ENSDART00000124223
|
ehmt1b
|
euchromatic histone-lysine N-methyltransferase 1b |
| chr15_+_36941490 | 1.20 |
ENSDART00000172664
|
kirrel3l
|
kirre like nephrin family adhesion molecule 3, like |
| chr5_+_13521081 | 1.20 |
ENSDART00000171975
|
si:ch211-230g14.6
|
si:ch211-230g14.6 |
| chr24_-_20599781 | 1.19 |
ENSDART00000179664
ENSDART00000141823 |
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr11_+_21586335 | 1.17 |
ENSDART00000091182
|
FAM72B
|
zgc:101564 |
| chr17_+_1499238 | 1.17 |
ENSDART00000187561
|
LO018430.1
|
|
| chr20_-_14781904 | 1.09 |
ENSDART00000187200
ENSDART00000179912 ENSDART00000160481 ENSDART00000026969 |
suco
|
SUN domain containing ossification factor |
| chr2_+_25019387 | 1.07 |
ENSDART00000142601
|
stag1a
|
stromal antigen 1a |
| chr20_+_40769265 | 1.06 |
ENSDART00000061168
|
tbc1d32
|
TBC1 domain family, member 32 |
| chr1_-_46401385 | 1.03 |
ENSDART00000150029
|
atp11a
|
ATPase phospholipid transporting 11A |
| chr19_-_17996162 | 1.01 |
ENSDART00000150928
ENSDART00000104491 |
ints8
|
integrator complex subunit 8 |
| chr19_+_40122160 | 1.01 |
ENSDART00000143966
|
si:ch211-173p18.3
|
si:ch211-173p18.3 |
| chr6_+_103361 | 0.98 |
ENSDART00000151899
|
ldlrb
|
low density lipoprotein receptor b |
| chr21_-_33995213 | 0.97 |
ENSDART00000140184
|
EBF1 (1 of many)
|
si:ch211-51e8.2 |
| chr15_+_47746176 | 0.91 |
ENSDART00000154481
ENSDART00000160914 |
stard10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr19_-_82504 | 0.88 |
ENSDART00000027864
ENSDART00000160560 |
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr14_-_21097574 | 0.88 |
ENSDART00000186803
|
rnf20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr25_-_27729046 | 0.84 |
ENSDART00000131437
|
zgc:153935
|
zgc:153935 |
| chr11_-_44999858 | 0.83 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr3_-_19561058 | 0.83 |
ENSDART00000079323
|
zgc:163079
|
zgc:163079 |
| chr8_+_15269423 | 0.81 |
ENSDART00000020386
|
gclm
|
glutamate-cysteine ligase, modifier subunit |
| chr25_-_1124851 | 0.78 |
ENSDART00000067558
|
spg11
|
spastic paraplegia 11 |
| chr2_-_58919975 | 0.78 |
ENSDART00000114286
|
sugp1
|
SURP and G patch domain containing 1 |
| chr8_+_39570615 | 0.72 |
ENSDART00000142557
|
lzts1
|
leucine zipper, putative tumor suppressor 1 |
| chr23_-_18982499 | 0.71 |
ENSDART00000012507
|
bcl2l1
|
bcl2-like 1 |
| chr23_+_9057999 | 0.70 |
ENSDART00000091899
|
ccm2l
|
cerebral cavernous malformation 2-like |
| chr23_+_7471072 | 0.69 |
ENSDART00000135551
|
si:ch211-200e2.1
|
si:ch211-200e2.1 |
| chr13_-_39947335 | 0.66 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr14_+_40852497 | 0.65 |
ENSDART00000128588
ENSDART00000166065 |
taf7
|
TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr18_+_49225552 | 0.65 |
ENSDART00000135026
|
si:ch211-136a13.1
|
si:ch211-136a13.1 |
| chr13_+_32740509 | 0.63 |
ENSDART00000076423
ENSDART00000160138 |
sobpa
|
sine oculis binding protein homolog (Drosophila) a |
| chr5_+_69697800 | 0.62 |
ENSDART00000178736
ENSDART00000162519 |
znf1005
|
zinc finger protein 1005 |
| chr24_-_10897511 | 0.62 |
ENSDART00000145593
ENSDART00000102484 ENSDART00000066784 |
fam49bb
|
family with sequence similarity 49, member Bb |
| chr17_-_23895026 | 0.58 |
ENSDART00000122108
|
pdzd8
|
PDZ domain containing 8 |
| chr10_-_4961923 | 0.57 |
ENSDART00000050177
ENSDART00000146066 |
snx30
|
sorting nexin family member 30 |
| chr5_-_61588998 | 0.56 |
ENSDART00000050912
|
pex12
|
peroxisomal biogenesis factor 12 |
| chr24_-_13566015 | 0.54 |
ENSDART00000123450
|
KCNB2
|
si:dkey-192j17.1 |
| chr5_-_68927728 | 0.52 |
ENSDART00000132838
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr5_+_31811662 | 0.51 |
ENSDART00000023463
|
uap1l1
|
UDP-N-acetylglucosamine pyrophosphorylase 1, like 1 |
| chr1_+_40613297 | 0.49 |
ENSDART00000040798
ENSDART00000168067 ENSDART00000130490 |
naa15b
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit b |
| chr3_-_37588855 | 0.49 |
ENSDART00000149258
|
arf2a
|
ADP-ribosylation factor 2a |
| chr7_+_27220655 | 0.48 |
ENSDART00000173987
ENSDART00000173583 |
plekha7b
|
pleckstrin homology domain containing, family A member 7b |
| chr8_-_7851590 | 0.44 |
ENSDART00000160692
|
zgc:113363
|
zgc:113363 |
| chr11_+_34921492 | 0.43 |
ENSDART00000128070
|
gnai2a
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2a |
| chr20_-_28642061 | 0.43 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr4_+_76349919 | 0.42 |
ENSDART00000174194
|
si:ch73-158p21.2
|
si:ch73-158p21.2 |
| chr13_+_13821054 | 0.41 |
ENSDART00000127417
|
slc4a11
|
solute carrier family 4, sodium borate transporter, member 11 |
| chr22_-_18116635 | 0.37 |
ENSDART00000005724
|
ncanb
|
neurocan b |
| chr8_+_12925385 | 0.37 |
ENSDART00000085377
|
KIF2A
|
zgc:103670 |
| chr19_-_81851 | 0.35 |
ENSDART00000172319
|
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr11_-_22605981 | 0.27 |
ENSDART00000186923
|
myog
|
myogenin |
| chr6_-_59942335 | 0.26 |
ENSDART00000168416
|
fbxl3b
|
F-box and leucine-rich repeat protein 3b |
| chr24_-_21587335 | 0.26 |
ENSDART00000091528
|
gpr12
|
G protein-coupled receptor 12 |
| chr5_+_30596822 | 0.26 |
ENSDART00000188375
|
hinfp
|
histone H4 transcription factor |
| chr6_-_7438584 | 0.25 |
ENSDART00000053776
|
fkbp11
|
FK506 binding protein 11 |
| chr10_+_26973063 | 0.25 |
ENSDART00000143162
ENSDART00000186210 |
tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr19_+_35799384 | 0.22 |
ENSDART00000076023
|
angpt2b
|
angiopoietin 2b |
| chr21_-_32289356 | 0.21 |
ENSDART00000183050
|
clk4b
|
CDC-like kinase 4b |
| chr10_+_10677697 | 0.21 |
ENSDART00000188705
|
fam163b
|
family with sequence similarity 163, member B |
| chr14_+_790166 | 0.19 |
ENSDART00000123912
|
adra2da
|
adrenergic, alpha-2D-, receptor a |
| chr5_-_67750907 | 0.18 |
ENSDART00000172097
|
b4galt4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr6_+_23026714 | 0.18 |
ENSDART00000124948
|
srp68
|
signal recognition particle 68 |
| chr1_-_17711361 | 0.16 |
ENSDART00000078848
|
ufsp2
|
ufm1-specific peptidase 2 |
| chr9_-_37748513 | 0.14 |
ENSDART00000188967
ENSDART00000187886 |
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
| chr1_-_17711636 | 0.14 |
ENSDART00000148322
ENSDART00000122670 |
ufsp2
|
ufm1-specific peptidase 2 |
| chr11_-_6070192 | 0.12 |
ENSDART00000162776
|
babam1
|
BRISC and BRCA1 A complex member 1 |
| chr17_-_4245902 | 0.10 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr17_-_14523722 | 0.10 |
ENSDART00000024726
|
daam1a
|
dishevelled associated activator of morphogenesis 1a |
| chr21_-_37973819 | 0.08 |
ENSDART00000133405
|
ripply1
|
ripply transcriptional repressor 1 |
| chr22_-_26558166 | 0.05 |
ENSDART00000111125
|
glis2a
|
GLIS family zinc finger 2a |
| chr8_-_1266181 | 0.04 |
ENSDART00000148654
ENSDART00000149924 |
cdc14b
|
cell division cycle 14B |
| chr25_+_2776511 | 0.03 |
ENSDART00000115280
|
neo1b
|
neogenin 1b |
Network of associatons between targets according to the STRING database.
First level regulatory network of zic4+zic6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.6 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.7 | 2.0 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.4 | 2.0 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.4 | 1.9 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.4 | 3.9 | GO:0034394 | protein localization to cell surface(GO:0034394) energy homeostasis(GO:0097009) |
| 0.3 | 3.5 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.3 | 1.3 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.2 | 0.7 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.2 | 0.8 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.2 | 3.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.2 | 1.3 | GO:2001270 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 2.8 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.2 | 1.3 | GO:0009146 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.2 | 3.8 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 2.8 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 5.7 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 2.2 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.1 | 0.6 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.1 | 1.1 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.1 | 1.6 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 3.0 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.1 | 0.7 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.1 | 2.3 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 0.5 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 1.5 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.2 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 0.7 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 4.9 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 3.0 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 1.4 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 2.9 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 1.4 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.5 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.4 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.2 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 3.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.8 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 1.0 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 1.1 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.6 | GO:1903039 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 1.1 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.1 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.1 | GO:0072401 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.0 | 1.2 | GO:0048846 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.0 | 0.8 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 0.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.9 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.3 | 2.8 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.2 | 0.6 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.2 | 2.9 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 2.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.6 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 5.5 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 2.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 2.0 | GO:0044327 | dendritic spine head(GO:0044327) phagocytic vesicle(GO:0045335) |
| 0.0 | 1.1 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 3.8 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.7 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 1.6 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 2.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 3.6 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 3.5 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 2.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 1.0 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 7.2 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.9 | GO:0031782 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.7 | 3.6 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.3 | 3.1 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.3 | 2.9 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 3.8 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 2.8 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.2 | 1.3 | GO:0016793 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) |
| 0.2 | 3.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.2 | 3.0 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.2 | 0.6 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.2 | 3.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 2.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 3.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 2.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 1.3 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.1 | 0.5 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 2.0 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 1.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.7 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 1.6 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 0.7 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 5.5 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.7 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.4 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 6.9 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 6.0 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 5.0 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 3.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.6 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.8 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 2.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 0.7 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 2.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 0.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 2.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 3.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.7 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 2.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |