Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
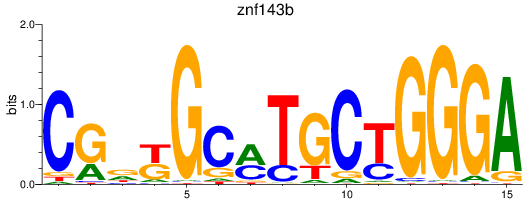
Results for znf143b
Z-value: 0.93
Transcription factors associated with znf143b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
znf143b
|
ENSDARG00000041581 | zinc finger protein 143b |
|
znf143b
|
ENSDARG00000111018 | zinc finger protein 143b |
|
znf143b
|
ENSDARG00000112408 | zinc finger protein 143b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| znf143b | dr11_v1_chr18_-_16937008_16937109 | 0.14 | 1.8e-01 | Click! |
Activity profile of znf143b motif
Sorted Z-values of znf143b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_50310684 | 13.03 |
ENSDART00000112152
|
gas7a
|
growth arrest-specific 7a |
| chr15_+_47161917 | 10.11 |
ENSDART00000167860
|
gap43
|
growth associated protein 43 |
| chr23_+_35714574 | 9.96 |
ENSDART00000164616
|
tuba1c
|
tubulin, alpha 1c |
| chr25_-_13842618 | 8.32 |
ENSDART00000160258
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr6_-_27057702 | 7.02 |
ENSDART00000149363
|
stk25a
|
serine/threonine kinase 25a |
| chr15_-_33933790 | 6.98 |
ENSDART00000165162
ENSDART00000182258 ENSDART00000183240 |
mag
|
myelin associated glycoprotein |
| chr4_-_1324141 | 6.73 |
ENSDART00000180720
|
ptn
|
pleiotrophin |
| chr22_+_12366516 | 6.31 |
ENSDART00000157802
|
r3hdm1
|
R3H domain containing 1 |
| chr8_-_43158486 | 6.30 |
ENSDART00000134801
|
ccdc92
|
coiled-coil domain containing 92 |
| chr21_-_16399778 | 6.25 |
ENSDART00000136435
|
unc5da
|
unc-5 netrin receptor Da |
| chr15_-_9257136 | 6.08 |
ENSDART00000187901
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr13_+_28819768 | 5.95 |
ENSDART00000191401
ENSDART00000188895 ENSDART00000101653 |
CU639469.1
|
|
| chr5_+_54685175 | 5.88 |
ENSDART00000115016
|
pmchl
|
pro-melanin-concentrating hormone, like |
| chr7_+_61050329 | 5.61 |
ENSDART00000115355
|
nwd2
|
NACHT and WD repeat domain containing 2 |
| chr6_-_48311 | 5.53 |
ENSDART00000131010
|
zgc:114175
|
zgc:114175 |
| chr4_+_25950372 | 5.31 |
ENSDART00000125767
|
metap2a
|
methionyl aminopeptidase 2a |
| chr18_+_41495841 | 5.31 |
ENSDART00000098671
|
si:ch211-203b8.6
|
si:ch211-203b8.6 |
| chr15_-_9419472 | 5.17 |
ENSDART00000122117
|
sacs
|
sacsin molecular chaperone |
| chr13_+_15816573 | 5.15 |
ENSDART00000137061
|
klc1a
|
kinesin light chain 1a |
| chr5_+_20148671 | 4.98 |
ENSDART00000143205
|
svopa
|
SV2 related protein a |
| chr15_-_12500938 | 4.97 |
ENSDART00000159627
|
scn4ba
|
sodium channel, voltage-gated, type IV, beta a |
| chr19_-_7358184 | 4.84 |
ENSDART00000092379
|
oxr1b
|
oxidation resistance 1b |
| chr20_-_16849306 | 4.80 |
ENSDART00000131395
ENSDART00000027582 |
brms1lb
|
breast cancer metastasis-suppressor 1-like b |
| chr17_-_22067451 | 4.66 |
ENSDART00000156872
|
ttbk1b
|
tau tubulin kinase 1b |
| chr24_+_15899365 | 4.55 |
ENSDART00000185965
|
neto1l
|
neuropilin (NRP) and tolloid (TLL)-like 1, like |
| chr14_+_25465346 | 4.52 |
ENSDART00000173436
|
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr19_-_1023051 | 4.49 |
ENSDART00000158429
|
tmem42b
|
transmembrane protein 42b |
| chr5_-_71722257 | 4.45 |
ENSDART00000013404
|
ak1
|
adenylate kinase 1 |
| chr9_-_27442339 | 4.40 |
ENSDART00000138602
|
stxbp5l
|
syntaxin binding protein 5-like |
| chr23_-_24825863 | 4.29 |
ENSDART00000112493
|
syt6a
|
synaptotagmin VIa |
| chr4_+_4232562 | 4.24 |
ENSDART00000177529
|
smkr1
|
small lysine rich protein 1 |
| chr10_-_27009413 | 4.06 |
ENSDART00000139942
ENSDART00000146983 ENSDART00000132352 |
uqcc3
|
ubiquinol-cytochrome c reductase complex assembly factor 3 |
| chr24_+_39129316 | 4.02 |
ENSDART00000155346
|
tbc1d24
|
TBC1 domain family, member 24 |
| chr3_-_50127668 | 3.94 |
ENSDART00000062116
|
cldnk
|
claudin k |
| chr5_+_65536095 | 3.90 |
ENSDART00000189898
|
si:dkey-21e5.1
|
si:dkey-21e5.1 |
| chr5_-_68641353 | 3.82 |
ENSDART00000048432
|
dlg4a
|
discs, large homolog 4a (Drosophila) |
| chr5_+_42467867 | 3.79 |
ENSDART00000172028
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr24_-_31843173 | 3.76 |
ENSDART00000185782
|
steap2
|
STEAP family member 2, metalloreductase |
| chr14_+_36521553 | 3.71 |
ENSDART00000136233
|
TENM3
|
si:dkey-237h12.3 |
| chr1_+_25848231 | 3.55 |
ENSDART00000027973
|
fabp2
|
fatty acid binding protein 2, intestinal |
| chr15_-_47193564 | 3.45 |
ENSDART00000172453
|
LSAMP
|
limbic system-associated membrane protein |
| chr9_+_26086135 | 3.36 |
ENSDART00000011910
|
arglu1a
|
arginine and glutamate rich 1a |
| chr16_-_8132742 | 3.34 |
ENSDART00000104323
|
snrka
|
SNF related kinase a |
| chr10_-_7858553 | 3.25 |
ENSDART00000182010
|
inpp5ja
|
inositol polyphosphate-5-phosphatase Ja |
| chr25_+_35706493 | 3.20 |
ENSDART00000176741
|
kif21a
|
kinesin family member 21A |
| chr9_-_7257877 | 3.16 |
ENSDART00000153514
|
si:ch211-121j5.4
|
si:ch211-121j5.4 |
| chr7_-_62003831 | 3.09 |
ENSDART00000113585
ENSDART00000062704 |
plaa
|
phospholipase A2-activating protein |
| chr17_+_25414033 | 3.08 |
ENSDART00000001691
|
tdh2
|
L-threonine dehydrogenase 2 |
| chr9_-_29985390 | 3.07 |
ENSDART00000134157
|
il1rapl1a
|
interleukin 1 receptor accessory protein-like 1a |
| chr6_+_53361874 | 3.05 |
ENSDART00000180660
|
si:ch211-161c3.5
|
si:ch211-161c3.5 |
| chr2_+_6127593 | 2.95 |
ENSDART00000184007
|
fzr1b
|
fizzy/cell division cycle 20 related 1b |
| chr2_-_12243213 | 2.94 |
ENSDART00000113081
|
gpr158b
|
G protein-coupled receptor 158b |
| chr18_+_28102620 | 2.91 |
ENSDART00000132342
|
kiaa1549lb
|
KIAA1549-like b |
| chr22_+_465269 | 2.91 |
ENSDART00000145767
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr22_+_797105 | 2.82 |
ENSDART00000128549
|
cry1bb
|
cryptochrome circadian clock 1bb |
| chr21_+_21791343 | 2.70 |
ENSDART00000151654
|
neu3.1
|
sialidase 3 (membrane sialidase), tandem duplicate 1 |
| chr25_-_20258508 | 2.68 |
ENSDART00000133860
ENSDART00000006840 ENSDART00000173434 |
dnm1l
|
dynamin 1-like |
| chr2_+_11670270 | 2.59 |
ENSDART00000100524
|
ftr01
|
finTRIM family, member 1 |
| chr16_+_46000956 | 2.51 |
ENSDART00000101753
ENSDART00000162393 |
mtmr11
|
myotubularin related protein 11 |
| chr20_-_35040041 | 2.50 |
ENSDART00000131919
|
kif26bb
|
kinesin family member 26Bb |
| chr11_-_41996957 | 2.50 |
ENSDART00000055706
|
her15.2
|
hairy and enhancer of split-related 15, tandem duplicate 2 |
| chr16_+_31511739 | 2.47 |
ENSDART00000049420
|
ndrg1b
|
N-myc downstream regulated 1b |
| chr4_+_12111154 | 2.45 |
ENSDART00000036779
|
tmem178b
|
transmembrane protein 178B |
| chr19_+_41701660 | 2.40 |
ENSDART00000033362
|
gatad2b
|
GATA zinc finger domain containing 2B |
| chr6_-_53282572 | 2.40 |
ENSDART00000172615
ENSDART00000165036 |
rbm5
|
RNA binding motif protein 5 |
| chr3_-_30861177 | 2.38 |
ENSDART00000154811
|
shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr14_-_31562795 | 2.38 |
ENSDART00000034979
|
mospd1
|
motile sperm domain containing 1 |
| chr13_-_49819027 | 2.37 |
ENSDART00000067824
|
b3galnt2
|
beta-1,3-N-acetylgalactosaminyltransferase 2 |
| chr21_+_7605803 | 2.35 |
ENSDART00000121813
|
wdr41
|
WD repeat domain 41 |
| chr2_-_5942115 | 2.34 |
ENSDART00000154489
|
tmem125b
|
transmembrane protein 125b |
| chr13_+_33055548 | 2.34 |
ENSDART00000137315
|
rdh12
|
retinol dehydrogenase 12 (all-trans/9-cis/11-cis) |
| chr15_-_766015 | 2.30 |
ENSDART00000190648
|
si:dkey-7i4.15
|
si:dkey-7i4.15 |
| chr3_-_33573252 | 2.29 |
ENSDART00000191339
ENSDART00000184612 ENSDART00000187232 ENSDART00000190671 |
CR847537.1
|
|
| chr4_-_75047192 | 2.29 |
ENSDART00000193576
|
large1
|
LARGE xylosyl- and glucuronyltransferase 1 |
| chr7_-_11605185 | 2.29 |
ENSDART00000169291
ENSDART00000113904 |
stard5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr9_+_28693592 | 2.27 |
ENSDART00000110198
|
zgc:162780
|
zgc:162780 |
| chr10_+_42678520 | 2.16 |
ENSDART00000182496
|
rhobtb2b
|
Rho-related BTB domain containing 2b |
| chr17_-_14671098 | 2.15 |
ENSDART00000037371
|
ppp1r13ba
|
protein phosphatase 1, regulatory subunit 13Ba |
| chr24_+_19591893 | 2.15 |
ENSDART00000152026
|
slco5a1a
|
solute carrier organic anion transporter family member 5A1a |
| chr10_-_43392267 | 2.11 |
ENSDART00000142872
|
edil3b
|
EGF-like repeats and discoidin I-like domains 3b |
| chr2_+_53357953 | 2.10 |
ENSDART00000187577
|
celf5b
|
cugbp, Elav-like family member 5b |
| chr25_+_8662469 | 2.09 |
ENSDART00000154680
ENSDART00000188568 |
man2a2
|
mannosidase, alpha, class 2A, member 2 |
| chr7_+_427503 | 2.07 |
ENSDART00000185942
|
NRXN2 (1 of many)
|
neurexin 2 |
| chr24_-_13364311 | 2.01 |
ENSDART00000183808
|
KCNB2
|
si:dkey-192j17.1 |
| chr5_-_62988463 | 1.99 |
ENSDART00000047143
|
specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr21_+_21791799 | 1.97 |
ENSDART00000151759
|
neu3.1
|
sialidase 3 (membrane sialidase), tandem duplicate 1 |
| chr22_+_696931 | 1.93 |
ENSDART00000149712
ENSDART00000009756 |
gpr37l1a
|
G protein-coupled receptor 37 like 1a |
| chr16_+_13822137 | 1.92 |
ENSDART00000163251
|
flcn
|
folliculin |
| chr24_+_19863246 | 1.91 |
ENSDART00000165242
|
CU638714.1
|
|
| chr14_+_25464681 | 1.91 |
ENSDART00000067500
ENSDART00000187601 |
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr15_-_11341635 | 1.89 |
ENSDART00000055220
|
rab30
|
RAB30, member RAS oncogene family |
| chr25_+_12012 | 1.88 |
ENSDART00000170348
|
cntn1a
|
contactin 1a |
| chr25_-_21609645 | 1.85 |
ENSDART00000087817
|
DOCK4 (1 of many)
|
si:dkey-81e3.1 |
| chr2_-_30912307 | 1.84 |
ENSDART00000188653
|
myl12.2
|
myosin, light chain 12, genome duplicate 2 |
| chr20_-_37629084 | 1.84 |
ENSDART00000141734
|
hivep2a
|
human immunodeficiency virus type I enhancer binding protein 2a |
| chr19_+_19032141 | 1.80 |
ENSDART00000192432
|
cpne4b
|
copine IVb |
| chr19_+_177455 | 1.80 |
ENSDART00000111580
|
tmem65
|
transmembrane protein 65 |
| chr6_-_37468971 | 1.80 |
ENSDART00000126379
|
plcxd1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
| chr7_+_50766094 | 1.79 |
ENSDART00000165037
|
si:ch73-380l10.2
|
si:ch73-380l10.2 |
| chr19_-_33996277 | 1.75 |
ENSDART00000159384
|
elmo1
|
engulfment and cell motility 1 (ced-12 homolog, C. elegans) |
| chr10_-_43392667 | 1.75 |
ENSDART00000183033
|
edil3b
|
EGF-like repeats and discoidin I-like domains 3b |
| chr8_+_19977712 | 1.74 |
ENSDART00000134124
|
znf692
|
zinc finger protein 692 |
| chr16_-_2558653 | 1.74 |
ENSDART00000110365
|
adcy3a
|
adenylate cyclase 3a |
| chr3_+_32477782 | 1.70 |
ENSDART00000151261
|
scaf1
|
SR-related CTD-associated factor 1 |
| chr23_-_4019928 | 1.68 |
ENSDART00000021062
|
slc9a8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr12_-_46959990 | 1.65 |
ENSDART00000084557
|
lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr15_+_21744680 | 1.64 |
ENSDART00000016860
|
ppp2r1bb
|
protein phosphatase 2, regulatory subunit A, beta b |
| chr5_+_59534286 | 1.63 |
ENSDART00000189341
|
gtf2ird1
|
GTF2I repeat domain containing 1 |
| chr14_+_41409697 | 1.54 |
ENSDART00000173335
|
bcorl1
|
BCL6 corepressor-like 1 |
| chr3_+_18050667 | 1.53 |
ENSDART00000035531
|
mettl26
|
methyltransferase like 26 |
| chr19_+_1835940 | 1.53 |
ENSDART00000167372
|
ptk2aa
|
protein tyrosine kinase 2aa |
| chr17_+_37253706 | 1.52 |
ENSDART00000076004
|
tmem62
|
transmembrane protein 62 |
| chr17_+_32571584 | 1.50 |
ENSDART00000087565
|
eva1a
|
eva-1 homolog A (C. elegans) |
| chr14_-_21661015 | 1.50 |
ENSDART00000189403
ENSDART00000172442 ENSDART00000181913 |
kdm3b
|
lysine (K)-specific demethylase 3B |
| chr10_-_8434816 | 1.49 |
ENSDART00000108643
|
tctn1
|
tectonic family member 1 |
| chr13_+_30506781 | 1.49 |
ENSDART00000110884
|
zmiz1a
|
zinc finger, MIZ-type containing 1a |
| chr17_-_14780578 | 1.48 |
ENSDART00000154690
|
si:ch211-266o15.1
|
si:ch211-266o15.1 |
| chr14_-_30876708 | 1.45 |
ENSDART00000147597
|
ubl3b
|
ubiquitin-like 3b |
| chr8_-_44904723 | 1.45 |
ENSDART00000040804
|
praf2
|
PRA1 domain family, member 2 |
| chr3_+_39568290 | 1.44 |
ENSDART00000020741
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr14_+_32852388 | 1.43 |
ENSDART00000166351
|
nkrf
|
NFKB repressing factor |
| chr1_-_55196103 | 1.40 |
ENSDART00000140153
|
mri1
|
methylthioribose-1-phosphate isomerase 1 |
| chr24_+_39576071 | 1.39 |
ENSDART00000147137
|
si:dkey-161j23.6
|
si:dkey-161j23.6 |
| chr10_+_31222656 | 1.35 |
ENSDART00000140988
ENSDART00000143387 |
tmem218
|
transmembrane protein 218 |
| chr3_-_40301467 | 1.34 |
ENSDART00000055186
|
atp5mf
|
ATP synthase membrane subunit f |
| chr12_-_48992527 | 1.32 |
ENSDART00000169696
|
CDHR1 (1 of many)
|
cadherin related family member 1 |
| chr5_-_23583446 | 1.30 |
ENSDART00000027217
|
cwc15
|
CWC15 spliceosome-associated protein homolog (S. cerevisiae) |
| chr21_-_43482426 | 1.29 |
ENSDART00000192901
|
ankrd46a
|
ankyrin repeat domain 46a |
| chr5_+_34549365 | 1.29 |
ENSDART00000009500
|
aif1l
|
allograft inflammatory factor 1-like |
| chr3_+_62236443 | 1.29 |
ENSDART00000180482
ENSDART00000193718 |
BX470259.4
|
|
| chr17_-_2126517 | 1.28 |
ENSDART00000013093
|
zfyve19
|
zinc finger, FYVE domain containing 19 |
| chr19_+_37458610 | 1.27 |
ENSDART00000103151
|
dlgap3
|
discs, large (Drosophila) homolog-associated protein 3 |
| chr6_-_50730749 | 1.25 |
ENSDART00000157153
ENSDART00000110441 |
pigu
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr22_-_2929127 | 1.24 |
ENSDART00000123730
ENSDART00000046678 |
pak2b
|
p21 protein (Cdc42/Rac)-activated kinase 2b |
| chr11_-_2222440 | 1.22 |
ENSDART00000162167
|
smug1
|
single-strand-selective monofunctional uracil-DNA glycosylase 1 |
| chr3_+_62219158 | 1.22 |
ENSDART00000162354
|
BX470259.5
|
|
| chr22_-_18779232 | 1.21 |
ENSDART00000186726
|
atp5f1d
|
ATP synthase F1 subunit delta |
| chr2_+_31437547 | 1.20 |
ENSDART00000141170
|
stam
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| chr1_+_26099250 | 1.20 |
ENSDART00000054205
|
ndufb6
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6 |
| chr7_+_67748939 | 1.17 |
ENSDART00000162978
|
dhx38
|
DEAH (Asp-Glu-Ala-His) box polypeptide 38 |
| chr18_+_36806545 | 1.16 |
ENSDART00000098958
|
ttc9b
|
tetratricopeptide repeat domain 9B |
| chr18_-_11060548 | 1.16 |
ENSDART00000146692
|
tspan9a
|
tetraspanin 9a |
| chr9_+_33158191 | 1.14 |
ENSDART00000180786
|
dopey2
|
dopey family member 2 |
| chr22_+_17531020 | 1.14 |
ENSDART00000137689
|
hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr15_+_5116179 | 1.14 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr15_+_44053244 | 1.12 |
ENSDART00000059550
|
lrrc51
|
leucine rich repeat containing 51 |
| chr23_-_4019699 | 1.11 |
ENSDART00000159780
|
slc9a8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr4_+_11483335 | 1.10 |
ENSDART00000184978
ENSDART00000140954 |
asb13a.2
|
ankyrin repeat and SOCS box containing 13a, tandem duplicate 2 |
| chr14_-_30876299 | 1.10 |
ENSDART00000180305
|
ubl3b
|
ubiquitin-like 3b |
| chr3_+_55031685 | 1.09 |
ENSDART00000132587
|
mpg
|
N-methylpurine DNA glycosylase |
| chr25_+_8925934 | 1.08 |
ENSDART00000073914
|
accs
|
1-aminocyclopropane-1-carboxylate synthase homolog (Arabidopsis)(non-functional) |
| chr2_-_1227221 | 1.07 |
ENSDART00000130897
|
ABCF3
|
ATP binding cassette subfamily F member 3 |
| chr22_-_506522 | 1.06 |
ENSDART00000106645
ENSDART00000067637 |
dstyk
|
dual serine/threonine and tyrosine protein kinase |
| chr5_+_34549845 | 1.05 |
ENSDART00000139317
|
aif1l
|
allograft inflammatory factor 1-like |
| chr15_-_1622468 | 1.04 |
ENSDART00000149008
ENSDART00000034456 |
kpna4
|
karyopherin alpha 4 (importin alpha 3) |
| chr7_-_56180680 | 1.04 |
ENSDART00000098438
|
spg7
|
spastic paraplegia 7 |
| chr23_+_31245395 | 1.03 |
ENSDART00000053588
|
irak1bp1
|
interleukin-1 receptor-associated kinase 1 binding protein 1 |
| chr7_+_62004048 | 1.01 |
ENSDART00000181818
|
smim20
|
small integral membrane protein 20 |
| chr16_+_12267672 | 1.00 |
ENSDART00000060037
|
zgc:92606
|
zgc:92606 |
| chr25_+_23358924 | 0.97 |
ENSDART00000156965
|
osbpl5
|
oxysterol binding protein-like 5 |
| chr11_+_5880562 | 0.96 |
ENSDART00000129663
ENSDART00000130768 ENSDART00000160909 |
dazap1
|
DAZ associated protein 1 |
| chr17_+_22760126 | 0.93 |
ENSDART00000151999
ENSDART00000190442 |
ttc27
|
tetratricopeptide repeat domain 27 |
| chr4_-_8058214 | 0.93 |
ENSDART00000132228
|
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr5_+_15495351 | 0.91 |
ENSDART00000111646
ENSDART00000114446 |
suds3
|
SDS3 homolog, SIN3A corepressor complex component |
| chr8_+_247163 | 0.90 |
ENSDART00000122378
|
cep120
|
centrosomal protein 120 |
| chr1_+_55095314 | 0.90 |
ENSDART00000132727
|
aftpha
|
aftiphilin a |
| chr2_+_42072689 | 0.89 |
ENSDART00000134203
|
vcpip1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr19_+_40069524 | 0.89 |
ENSDART00000151365
ENSDART00000140926 |
zmym4
|
zinc finger, MYM-type 4 |
| chr22_+_20172018 | 0.89 |
ENSDART00000188104
|
hmg20b
|
high mobility group 20B |
| chr1_+_44906517 | 0.87 |
ENSDART00000142820
|
wu:fc21g02
|
wu:fc21g02 |
| chr5_-_24642026 | 0.85 |
ENSDART00000139522
|
si:ch211-106a19.1
|
si:ch211-106a19.1 |
| chr20_-_9428021 | 0.85 |
ENSDART00000025330
|
rdh14b
|
retinol dehydrogenase 14b |
| chr16_-_39900665 | 0.84 |
ENSDART00000136719
|
rbms3
|
RNA binding motif, single stranded interacting protein |
| chr22_-_18778988 | 0.84 |
ENSDART00000019235
|
atp5f1d
|
ATP synthase F1 subunit delta |
| chr19_+_15521997 | 0.84 |
ENSDART00000003164
|
ppp1r8a
|
protein phosphatase 1, regulatory subunit 8a |
| chr13_-_37127970 | 0.83 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr21_-_13493608 | 0.81 |
ENSDART00000192307
|
nsmfa
|
NMDA receptor synaptonuclear signaling and neuronal migration factor a |
| chr23_+_37185247 | 0.80 |
ENSDART00000146269
|
vwa5b1
|
von Willebrand factor A domain containing 5B1 |
| chr9_-_55946377 | 0.77 |
ENSDART00000161536
ENSDART00000169432 |
SH3RF3
|
si:ch211-124n19.2 |
| chr14_+_989733 | 0.77 |
ENSDART00000161487
ENSDART00000127317 |
si:ch73-308l14.2
|
si:ch73-308l14.2 |
| chr5_+_13353866 | 0.77 |
ENSDART00000099611
|
ccl19a.1
|
chemokine (C-C motif) ligand 19a, tandem duplicate 1 |
| chr19_-_12967986 | 0.77 |
ENSDART00000151064
|
slc25a32a
|
solute carrier family 25 (mitochondrial folate carrier), member 32a |
| chr3_-_50066499 | 0.76 |
ENSDART00000056618
ENSDART00000154561 |
mrpl12
|
mitochondrial ribosomal protein L12 |
| chr16_-_34212912 | 0.75 |
ENSDART00000145017
|
phactr4b
|
phosphatase and actin regulator 4b |
| chr17_+_5846202 | 0.74 |
ENSDART00000189713
|
fndc4b
|
fibronectin type III domain containing 4b |
| chr22_+_17531214 | 0.71 |
ENSDART00000161174
ENSDART00000088356 |
hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr21_+_7131970 | 0.71 |
ENSDART00000161921
|
zgc:113019
|
zgc:113019 |
| chr13_+_31133270 | 0.69 |
ENSDART00000014922
|
arhgap22
|
Rho GTPase activating protein 22 |
| chr9_+_38624478 | 0.68 |
ENSDART00000077389
ENSDART00000114728 |
znf148
|
zinc finger protein 148 |
| chr10_+_37182626 | 0.68 |
ENSDART00000137636
|
ksr1a
|
kinase suppressor of ras 1a |
| chr21_-_23046606 | 0.66 |
ENSDART00000016167
|
zw10
|
zw10 kinetochore protein |
| chr15_-_39848822 | 0.65 |
ENSDART00000155230
|
si:dkey-263j23.1
|
si:dkey-263j23.1 |
| chr7_+_67749251 | 0.64 |
ENSDART00000167562
|
dhx38
|
DEAH (Asp-Glu-Ala-His) box polypeptide 38 |
| chr6_+_12482599 | 0.63 |
ENSDART00000090316
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr5_-_36949476 | 0.63 |
ENSDART00000047269
|
h3f3c
|
H3 histone, family 3C |
| chr15_-_47857687 | 0.62 |
ENSDART00000098982
ENSDART00000151594 |
h3f3b.1
|
H3 histone, family 3B.1 |
| chr23_-_45318760 | 0.61 |
ENSDART00000166883
|
ccdc171
|
coiled-coil domain containing 171 |
| chr1_+_53279861 | 0.61 |
ENSDART00000035713
|
rnf150a
|
ring finger protein 150a |
| chr10_+_26689262 | 0.61 |
ENSDART00000123195
ENSDART00000132946 |
tmtops3b
|
teleost multiple tissue opsin 3b |
| chr17_+_31914877 | 0.60 |
ENSDART00000177801
|
FAM196A (1 of many)
|
family with sequence similarity 196 member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of znf143b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.3 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 1.2 | 3.5 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.9 | 3.8 | GO:0015677 | copper ion import(GO:0015677) |
| 0.7 | 4.1 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.6 | 4.4 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.6 | 10.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.6 | 1.8 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.6 | 6.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.5 | 2.7 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.5 | 2.9 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.5 | 1.4 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.5 | 1.9 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.4 | 3.5 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.4 | 1.3 | GO:0009838 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.4 | 2.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.4 | 4.7 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.4 | 3.1 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.3 | 1.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.3 | 1.9 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.3 | 2.3 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.2 | 1.5 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.2 | 0.9 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.2 | 2.9 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.2 | 5.4 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.2 | 1.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.2 | 3.1 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.2 | 2.5 | GO:0045580 | regulation of T cell differentiation(GO:0045580) |
| 0.2 | 0.5 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 1.5 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 1.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 2.0 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 8.3 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.1 | 4.3 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 1.1 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.1 | 1.5 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 2.3 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.1 | 0.5 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.1 | 6.7 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.1 | 7.0 | GO:0003401 | axis elongation(GO:0003401) |
| 0.1 | 2.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.5 | GO:0017003 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.1 | 4.0 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.1 | 1.5 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 2.1 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 5.7 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 2.4 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 | 0.8 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 1.0 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.1 | 0.5 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.6 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.3 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 1.3 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 1.0 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 3.9 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 2.4 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.1 | 1.0 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 2.1 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 1.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.3 | GO:0043111 | replication fork arrest(GO:0043111) |
| 0.1 | 1.4 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 6.1 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 1.0 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 2.2 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.1 | 2.3 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 1.9 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 1.0 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.1 | 0.9 | GO:1903286 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 13.0 | GO:0030041 | actin filament polymerization(GO:0030041) |
| 0.1 | 1.0 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 5.9 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 4.3 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) |
| 0.1 | 2.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 2.8 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.6 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 1.8 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 2.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.8 | GO:0035308 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 1.8 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 2.8 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.3 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 1.9 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.7 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.0 | 0.8 | GO:1903844 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.2 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.8 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.4 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 1.6 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 9.5 | GO:0000278 | mitotic cell cycle(GO:0000278) |
| 0.0 | 0.5 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
| 0.0 | 3.3 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 1.5 | GO:0006914 | autophagy(GO:0006914) |
| 0.0 | 0.6 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 1.0 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 10.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.5 | 2.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.4 | 4.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.3 | 5.7 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.2 | 1.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.2 | 1.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 0.5 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 17.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 1.3 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 0.9 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 2.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.9 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 10.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 3.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 2.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.3 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 1.3 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 0.2 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 0.3 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.1 | 0.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.8 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 4.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 3.5 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 2.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.2 | GO:0036452 | ESCRT complex(GO:0036452) |
| 0.0 | 1.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.3 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 3.5 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 8.3 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 4.3 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 1.0 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 3.5 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 3.8 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 2.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 6.0 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.8 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 2.1 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.7 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 1.0 | GO:0099503 | secretory vesicle(GO:0099503) |
| 0.0 | 0.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.5 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.5 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 4.3 | GO:0097708 | cytoplasmic vesicle(GO:0031410) intracellular vesicle(GO:0097708) |
| 0.0 | 1.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 1.0 | 3.1 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.9 | 3.8 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.9 | 13.1 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.6 | 6.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.6 | 3.1 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.6 | 2.9 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.4 | 3.5 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.4 | 4.7 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.4 | 1.7 | GO:0101006 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.4 | 1.2 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.4 | 7.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.4 | 2.3 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.3 | 5.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.3 | 4.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.3 | 3.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.3 | 4.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.3 | 0.8 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.3 | 2.8 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 1.0 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.2 | 2.3 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.2 | 1.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 5.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 8.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 5.9 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 2.1 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 0.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 2.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 1.0 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 5.7 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 2.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 1.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.5 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.1 | 1.1 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 1.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 4.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.8 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 2.5 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.1 | 0.3 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 3.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 1.1 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.1 | 1.8 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 1.0 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 0.8 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.9 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 1.0 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 1.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 4.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.8 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.6 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 13.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 3.1 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.6 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.3 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 1.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 4.0 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 1.8 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 1.6 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 1.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.8 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 6.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 17.4 | GO:0004672 | protein kinase activity(GO:0004672) |
| 0.0 | 0.5 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.6 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 3.7 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.5 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 9.3 | GO:0005509 | calcium ion binding(GO:0005509) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.3 | 10.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 2.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 7.0 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 1.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 2.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.5 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 10.0 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.3 | 6.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 4.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 2.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 1.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.7 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.1 | 1.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 2.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.8 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 1.6 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 1.0 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 2.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |