Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
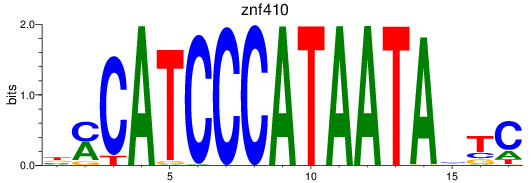
Results for znf410
Z-value: 0.46
Transcription factors associated with znf410
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
znf410
|
ENSDARG00000008218 | zinc finger protein 410 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| znf410 | dr11_v1_chr20_+_54349333_54349461 | -0.35 | 5.0e-04 | Click! |
Activity profile of znf410 motif
Sorted Z-values of znf410 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_31791165 | 12.19 |
ENSDART00000148389
|
chd4b
|
chromodomain helicase DNA binding protein 4b |
| chr19_-_5058908 | 10.85 |
ENSDART00000130062
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| chr3_+_18567560 | 2.34 |
ENSDART00000124329
|
cbx8a
|
chromobox homolog 8a (Pc class homolog, Drosophila) |
| chr6_+_18359306 | 1.82 |
ENSDART00000157796
|
cbx8b
|
chromobox homolog 8b |
| chr19_-_5058515 | 1.59 |
ENSDART00000150980
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| chr19_-_977849 | 1.51 |
ENSDART00000172303
|
CABZ01088282.1
|
|
| chr13_-_11800281 | 1.21 |
ENSDART00000079392
|
zmp:0000000662
|
zmp:0000000662 |
| chr10_-_94184 | 1.11 |
ENSDART00000146125
|
ttc3
|
tetratricopeptide repeat domain 3 |
| chr10_-_35186310 | 1.05 |
ENSDART00000127805
|
pom121
|
POM121 transmembrane nucleoporin |
| chr19_+_43396861 | 0.92 |
ENSDART00000151433
|
yrk
|
Yes-related kinase |
| chr8_-_12867434 | 0.92 |
ENSDART00000081657
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr4_+_76336370 | 0.76 |
ENSDART00000170520
|
zgc:171673
|
zgc:171673 |
Network of associatons between targets according to the STRING database.
First level regulatory network of znf410
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 12.4 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.9 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 1.0 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 0.0 | 1.8 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 12.4 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.0 | GO:0005643 | nuclear pore(GO:0005643) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 12.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 12.2 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.1 | 0.9 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 1.0 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.0 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |