Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
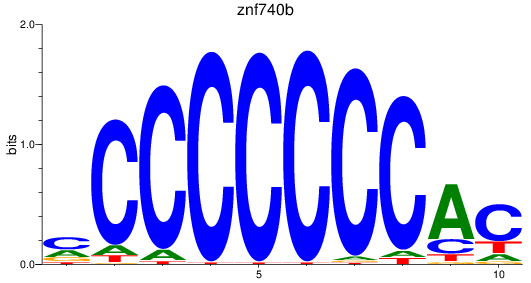
Results for znf740b
Z-value: 0.83
Transcription factors associated with znf740b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
znf740b
|
ENSDARG00000061174 | zinc finger protein 740b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| znf740b | dr11_v1_chr23_-_36418708_36418708 | 0.47 | 1.5e-06 | Click! |
Activity profile of znf740b motif
Sorted Z-values of znf740b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_36803415 | 8.20 |
ENSDART00000111680
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr25_+_4750972 | 7.59 |
ENSDART00000168903
|
si:zfos-2372e4.1
|
si:zfos-2372e4.1 |
| chr11_-_3334248 | 7.09 |
ENSDART00000154314
ENSDART00000121861 |
prph
|
peripherin |
| chr11_-_24191928 | 6.24 |
ENSDART00000136827
|
sox12
|
SRY (sex determining region Y)-box 12 |
| chr2_+_22694382 | 5.20 |
ENSDART00000139196
|
kif1ab
|
kinesin family member 1Ab |
| chr1_+_39553040 | 5.02 |
ENSDART00000137676
|
tenm3
|
teneurin transmembrane protein 3 |
| chr25_+_30196039 | 4.87 |
ENSDART00000005299
|
hsd17b12a
|
hydroxysteroid (17-beta) dehydrogenase 12a |
| chr3_-_61162750 | 4.49 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr25_+_37366698 | 4.39 |
ENSDART00000165400
ENSDART00000192589 |
slc1a2b
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2b |
| chr24_+_41931585 | 4.19 |
ENSDART00000130310
|
epb41l3a
|
erythrocyte membrane protein band 4.1-like 3a |
| chr10_+_1849874 | 4.01 |
ENSDART00000158897
ENSDART00000149956 |
apc
|
adenomatous polyposis coli |
| chr5_+_31214341 | 4.00 |
ENSDART00000133432
|
atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr1_-_42778510 | 3.99 |
ENSDART00000190172
|
lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr11_-_97817 | 3.94 |
ENSDART00000092903
|
elmo2
|
engulfment and cell motility 2 |
| chr21_+_13389499 | 3.91 |
ENSDART00000151268
|
zgc:113162
|
zgc:113162 |
| chr23_-_10175898 | 3.58 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr20_+_26538137 | 3.54 |
ENSDART00000045397
|
stx11b.1
|
syntaxin 11b, tandem duplicate 1 |
| chr16_-_17586883 | 3.48 |
ENSDART00000017142
|
m6pr
|
mannose-6-phosphate receptor (cation dependent) |
| chr22_-_20403194 | 3.45 |
ENSDART00000010048
|
map2k2a
|
mitogen-activated protein kinase kinase 2a |
| chr23_+_6232895 | 3.43 |
ENSDART00000139795
|
syt2a
|
synaptotagmin IIa |
| chr1_+_31112436 | 3.41 |
ENSDART00000075340
|
eef1a1b
|
eukaryotic translation elongation factor 1 alpha 1b |
| chr6_+_58492201 | 3.40 |
ENSDART00000156375
|
kcnq2b
|
potassium voltage-gated channel, KQT-like subfamily, member 2b |
| chr16_-_31435020 | 3.35 |
ENSDART00000138508
|
zgc:194210
|
zgc:194210 |
| chr6_+_38381957 | 3.30 |
ENSDART00000087300
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr20_+_20672163 | 3.30 |
ENSDART00000027758
|
rtn1b
|
reticulon 1b |
| chr20_-_25542183 | 3.30 |
ENSDART00000024350
|
cyp2ad2
|
cytochrome P450, family 2, subfamily AD, polypeptide 2 |
| chr17_-_12389259 | 3.24 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr12_+_24561832 | 3.19 |
ENSDART00000088159
|
nrxn1a
|
neurexin 1a |
| chr3_-_50998577 | 3.17 |
ENSDART00000157735
|
cdc42ep4a
|
CDC42 effector protein (Rho GTPase binding) 4a |
| chr7_-_27686021 | 3.11 |
ENSDART00000079112
ENSDART00000100989 |
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr2_-_16565690 | 3.04 |
ENSDART00000022549
|
atp1b3a
|
ATPase Na+/K+ transporting subunit beta 3a |
| chr21_+_1143141 | 3.00 |
ENSDART00000178294
|
CABZ01086139.1
|
|
| chr14_+_22172047 | 2.97 |
ENSDART00000114750
ENSDART00000148259 |
gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr10_-_17170086 | 2.95 |
ENSDART00000020122
|
ywhah
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide |
| chr16_-_25393699 | 2.92 |
ENSDART00000149445
|
pard6ga
|
par-6 family cell polarity regulator gamma a |
| chr13_-_27767330 | 2.84 |
ENSDART00000131631
ENSDART00000112553 ENSDART00000189911 |
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr11_+_25101220 | 2.80 |
ENSDART00000183700
|
ndrg3a
|
ndrg family member 3a |
| chr5_+_32162684 | 2.78 |
ENSDART00000134472
|
taok3b
|
TAO kinase 3b |
| chr14_+_31566517 | 2.72 |
ENSDART00000002684
|
ints6l
|
integrator complex subunit 6 like |
| chr11_+_38280454 | 2.70 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr13_+_11439486 | 2.70 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr1_-_42779075 | 2.69 |
ENSDART00000133917
|
lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr15_-_28618502 | 2.67 |
ENSDART00000086902
|
slc6a4a
|
solute carrier family 6 (neurotransmitter transporter), member 4a |
| chr3_+_21189766 | 2.64 |
ENSDART00000078807
|
zgc:123295
|
zgc:123295 |
| chr5_-_14326959 | 2.61 |
ENSDART00000137355
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr18_+_35742838 | 2.59 |
ENSDART00000088504
ENSDART00000140386 |
rasgrp4
|
RAS guanyl releasing protein 4 |
| chr25_+_4091067 | 2.54 |
ENSDART00000104926
|
eps8l2
|
EPS8 like 2 |
| chr21_+_22878834 | 2.50 |
ENSDART00000065562
|
pcf11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr8_-_31107537 | 2.46 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| chr13_-_638485 | 2.44 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr9_-_31278048 | 2.41 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr9_+_29585943 | 2.40 |
ENSDART00000185989
ENSDART00000115290 |
mcf2lb
|
mcf.2 cell line derived transforming sequence-like b |
| chr19_+_13994563 | 2.39 |
ENSDART00000164696
|
tmem222b
|
transmembrane protein 222b |
| chr17_+_37310663 | 2.31 |
ENSDART00000157122
|
elmsan1b
|
ELM2 and Myb/SANT-like domain containing 1b |
| chr21_+_30351256 | 2.31 |
ENSDART00000078341
|
foxi3a
|
forkhead box I3a |
| chr25_-_11088839 | 2.30 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr22_+_24157807 | 2.29 |
ENSDART00000159165
|
b3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr3_-_31804481 | 2.28 |
ENSDART00000028270
|
gfap
|
glial fibrillary acidic protein |
| chr21_-_9446747 | 2.28 |
ENSDART00000158790
|
ptpn13
|
protein tyrosine phosphatase, non-receptor type 13 |
| chr23_+_5977965 | 2.23 |
ENSDART00000115403
ENSDART00000183147 |
NAV1 (1 of many)
|
neuron navigator 1 |
| chr12_+_42436328 | 2.23 |
ENSDART00000167324
|
ebf3a
|
early B cell factor 3a |
| chr10_-_35542071 | 2.22 |
ENSDART00000162139
|
si:ch211-244c8.4
|
si:ch211-244c8.4 |
| chr16_-_47245563 | 2.21 |
ENSDART00000190225
|
LO017877.2
|
|
| chr23_+_39346930 | 2.12 |
ENSDART00000102843
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr3_+_13440900 | 2.10 |
ENSDART00000143715
|
GNG14
|
si:dkey-117i10.1 |
| chr9_-_32177117 | 2.09 |
ENSDART00000078568
|
sf3b1
|
splicing factor 3b, subunit 1 |
| chr16_-_9879407 | 2.07 |
ENSDART00000149424
|
ncalda
|
neurocalcin delta a |
| chr8_+_25267903 | 2.06 |
ENSDART00000093090
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr16_+_1803462 | 2.06 |
ENSDART00000183974
|
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr20_-_35246150 | 2.05 |
ENSDART00000090549
|
fzd3a
|
frizzled class receptor 3a |
| chr21_+_7823146 | 2.04 |
ENSDART00000030579
|
crhbp
|
corticotropin releasing hormone binding protein |
| chr16_-_24672919 | 2.02 |
ENSDART00000008326
|
pon2
|
paraoxonase 2 |
| chr8_+_28900689 | 1.98 |
ENSDART00000141634
|
grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr8_-_23776399 | 1.97 |
ENSDART00000114800
|
INAVA
|
si:ch211-163l21.4 |
| chr9_-_18877597 | 1.97 |
ENSDART00000099446
|
kctd4
|
potassium channel tetramerization domain containing 4 |
| chr17_+_52822422 | 1.96 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr5_+_36614196 | 1.95 |
ENSDART00000150574
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr3_-_26204867 | 1.94 |
ENSDART00000103748
|
gdpd3a
|
glycerophosphodiester phosphodiesterase domain containing 3a |
| chr22_-_5839686 | 1.92 |
ENSDART00000145406
|
si:rp71-36a1.5
|
si:rp71-36a1.5 |
| chr15_-_35246742 | 1.90 |
ENSDART00000131479
|
mff
|
mitochondrial fission factor |
| chr21_+_11244068 | 1.88 |
ENSDART00000163432
|
arid6
|
AT-rich interaction domain 6 |
| chr2_+_37245382 | 1.88 |
ENSDART00000004626
|
sec62
|
SEC62 homolog, preprotein translocation factor |
| chr17_+_5846202 | 1.87 |
ENSDART00000189713
|
fndc4b
|
fibronectin type III domain containing 4b |
| chr1_-_8012476 | 1.87 |
ENSDART00000177051
|
CR855320.3
|
|
| chr7_+_23495986 | 1.87 |
ENSDART00000190739
ENSDART00000115299 ENSDART00000101423 ENSDART00000142401 |
zgc:109889
|
zgc:109889 |
| chr11_-_4235811 | 1.87 |
ENSDART00000121716
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr9_+_7358749 | 1.87 |
ENSDART00000081660
|
ihha
|
Indian hedgehog homolog a |
| chr16_-_27749172 | 1.87 |
ENSDART00000145198
|
steap4
|
STEAP family member 4 |
| chr9_+_13714379 | 1.86 |
ENSDART00000017593
ENSDART00000145503 |
tmem237a
|
transmembrane protein 237a |
| chr13_-_45523026 | 1.85 |
ENSDART00000020663
|
rhd
|
Rh blood group, D antigen |
| chr13_+_23677949 | 1.84 |
ENSDART00000144215
|
pcnxl2
|
pecanex-like 2 (Drosophila) |
| chr12_-_20584413 | 1.82 |
ENSDART00000170923
|
FP885542.2
|
|
| chr6_-_29377092 | 1.82 |
ENSDART00000078665
|
tmem131
|
transmembrane protein 131 |
| chr9_-_32753535 | 1.81 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr5_-_67115872 | 1.79 |
ENSDART00000065262
|
rps6ka4
|
ribosomal protein S6 kinase, polypeptide 4 |
| chr12_-_14211293 | 1.79 |
ENSDART00000158399
|
avl9
|
AVL9 homolog (S. cerevisiase) |
| chr5_-_35264517 | 1.79 |
ENSDART00000114981
|
ptcd2
|
pentatricopeptide repeat domain 2 |
| chr9_-_23807032 | 1.78 |
ENSDART00000027443
|
esyt3
|
extended synaptotagmin-like protein 3 |
| chr18_+_21122818 | 1.78 |
ENSDART00000060015
ENSDART00000060184 |
chka
|
choline kinase alpha |
| chr15_+_3790556 | 1.77 |
ENSDART00000181467
|
RNF14 (1 of many)
|
ring finger protein 14 |
| chr8_+_36270571 | 1.75 |
ENSDART00000187505
|
CU929676.1
|
|
| chr20_-_37629084 | 1.74 |
ENSDART00000141734
|
hivep2a
|
human immunodeficiency virus type I enhancer binding protein 2a |
| chr2_-_23004286 | 1.74 |
ENSDART00000134664
ENSDART00000110373 ENSDART00000185833 ENSDART00000187235 |
znf414
mllt1b
|
zinc finger protein 414 MLLT1, super elongation complex subunit b |
| chr10_-_22249444 | 1.73 |
ENSDART00000148831
|
fgf11b
|
fibroblast growth factor 11b |
| chr20_-_20610812 | 1.73 |
ENSDART00000181870
|
ppm1ab
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Ab |
| chr2_+_58221163 | 1.72 |
ENSDART00000157939
|
FO704813.1
|
|
| chr9_+_8942258 | 1.69 |
ENSDART00000138836
|
ankrd10b
|
ankyrin repeat domain 10b |
| chr19_+_2602903 | 1.68 |
ENSDART00000033132
|
fam126a
|
family with sequence similarity 126, member A |
| chr1_-_38709551 | 1.66 |
ENSDART00000128794
|
gpm6ab
|
glycoprotein M6Ab |
| chr21_-_30181732 | 1.63 |
ENSDART00000015636
|
hnrnph1l
|
heterogeneous nuclear ribonucleoprotein H1, like |
| chr15_-_37543839 | 1.56 |
ENSDART00000148870
|
kmt2bb
|
lysine (K)-specific methyltransferase 2Bb |
| chr14_-_2369849 | 1.56 |
ENSDART00000180422
ENSDART00000189731 ENSDART00000111748 |
pcdhb
|
protocadherin b |
| chr7_-_2039060 | 1.56 |
ENSDART00000173879
|
si:cabz01007794.1
|
si:cabz01007794.1 |
| chr2_+_43469241 | 1.56 |
ENSDART00000142078
ENSDART00000098265 |
nrp1b
|
neuropilin 1b |
| chr5_+_42912966 | 1.55 |
ENSDART00000039973
|
rufy3
|
RUN and FYVE domain containing 3 |
| chr13_+_2830574 | 1.54 |
ENSDART00000138143
|
si:ch211-233m11.1
|
si:ch211-233m11.1 |
| chr5_-_70625697 | 1.52 |
ENSDART00000179538
ENSDART00000190540 |
CABZ01075572.1
|
|
| chr7_-_19043401 | 1.51 |
ENSDART00000158796
|
fcer1g
|
Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide |
| chr18_+_46151505 | 1.51 |
ENSDART00000015034
ENSDART00000141287 |
blvrb
|
biliverdin reductase B |
| chr6_+_38845697 | 1.50 |
ENSDART00000053187
|
stk35l
|
serine/threonine kinase 35, like |
| chr18_+_41232719 | 1.49 |
ENSDART00000138552
ENSDART00000145863 |
trip12
|
thyroid hormone receptor interactor 12 |
| chr25_+_35502552 | 1.49 |
ENSDART00000189612
ENSDART00000058443 |
fibina
|
fin bud initiation factor a |
| chr14_+_33458294 | 1.46 |
ENSDART00000075278
|
atp1b4
|
ATPase Na+/K+ transporting subunit beta 4 |
| chr2_+_47623202 | 1.46 |
ENSDART00000154465
|
si:ch211-165b10.3
|
si:ch211-165b10.3 |
| chr2_-_36818132 | 1.45 |
ENSDART00000110447
|
slitrk3b
|
SLIT and NTRK-like family, member 3b |
| chr11_+_6650966 | 1.45 |
ENSDART00000131236
|
si:dkey-246j7.1
|
si:dkey-246j7.1 |
| chr6_-_30859656 | 1.44 |
ENSDART00000156235
|
pde4ba
|
phosphodiesterase 4B, cAMP-specific a |
| chr25_-_19395476 | 1.44 |
ENSDART00000182622
|
map1ab
|
microtubule-associated protein 1Ab |
| chr2_+_28889936 | 1.42 |
ENSDART00000078232
|
cdh10a
|
cadherin 10, type 2a (T2-cadherin) |
| chr3_-_33417826 | 1.41 |
ENSDART00000084284
|
abi3a
|
ABI family, member 3a |
| chr11_-_23322182 | 1.40 |
ENSDART00000111289
|
kiss1
|
KiSS-1 metastasis-suppressor |
| chr15_-_37543591 | 1.39 |
ENSDART00000180400
|
kmt2bb
|
lysine (K)-specific methyltransferase 2Bb |
| chr3_+_37824268 | 1.39 |
ENSDART00000137038
|
asic2
|
acid-sensing (proton-gated) ion channel 2 |
| chr4_+_34417403 | 1.38 |
ENSDART00000186032
ENSDART00000167909 |
si:ch211-246b8.2
|
si:ch211-246b8.2 |
| chr19_+_12406583 | 1.36 |
ENSDART00000013865
ENSDART00000151535 |
seh1l
|
SEH1-like (S. cerevisiae) |
| chr22_-_5918670 | 1.35 |
ENSDART00000141373
|
si:rp71-36a1.1
|
si:rp71-36a1.1 |
| chr25_+_3788074 | 1.34 |
ENSDART00000154008
|
chid1
|
chitinase domain containing 1 |
| chr10_-_43721530 | 1.34 |
ENSDART00000025366
|
cetn3
|
centrin 3 |
| chr17_-_36988455 | 1.33 |
ENSDART00000187180
ENSDART00000126823 |
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr13_+_771403 | 1.32 |
ENSDART00000093166
|
nrxn1b
|
neurexin 1b |
| chr1_+_53842344 | 1.30 |
ENSDART00000114506
|
crfb17
|
cytokine receptor family member B17 |
| chr3_+_62126981 | 1.29 |
ENSDART00000060527
|
drc3
|
dynein regulatory complex subunit 3 |
| chr5_-_40190949 | 1.29 |
ENSDART00000175588
|
wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr2_-_37043540 | 1.28 |
ENSDART00000131834
|
gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr7_-_28611145 | 1.28 |
ENSDART00000054366
|
scube2
|
signal peptide, CUB domain, EGF-like 2 |
| chr11_-_7139555 | 1.27 |
ENSDART00000016472
|
bmp7a
|
bone morphogenetic protein 7a |
| chr12_+_8003872 | 1.27 |
ENSDART00000180300
ENSDART00000126661 |
rhobtb1
|
Rho-related BTB domain containing 1 |
| chr6_-_18121075 | 1.25 |
ENSDART00000171072
|
SEC14L1
|
si:dkey-237i9.1 |
| chr22_+_35068046 | 1.25 |
ENSDART00000161660
ENSDART00000169573 |
si:ch73-173h19.3
|
si:ch73-173h19.3 |
| chr11_+_7432533 | 1.24 |
ENSDART00000180977
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr15_+_32643873 | 1.23 |
ENSDART00000189433
|
trpc4b
|
transient receptor potential cation channel, subfamily C, member 4b |
| chr19_-_7358184 | 1.23 |
ENSDART00000092379
|
oxr1b
|
oxidation resistance 1b |
| chr16_-_41439659 | 1.22 |
ENSDART00000191624
|
cpne4a
|
copine IVa |
| chr23_-_524780 | 1.22 |
ENSDART00000055139
|
col9a3
|
collagen, type IX, alpha 3 |
| chr14_-_24111292 | 1.22 |
ENSDART00000186611
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr2_-_44183613 | 1.21 |
ENSDART00000079596
|
cadm3
|
cell adhesion molecule 3 |
| chr8_-_37328112 | 1.20 |
ENSDART00000140365
|
slc12a5b
|
solute carrier family 12 (potassium/chloride transporter), member 5b |
| chr8_-_23578660 | 1.20 |
ENSDART00000039080
|
wasb
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia) b |
| chr6_-_34860574 | 1.18 |
ENSDART00000073957
|
slc35d1a
|
solute carrier family 35 (UDP-GlcA/UDP-GalNAc transporter), member D1a |
| chr7_+_16835218 | 1.18 |
ENSDART00000173580
|
nav2a
|
neuron navigator 2a |
| chr12_-_22540943 | 1.15 |
ENSDART00000172310
|
zbtb4
|
zinc finger and BTB domain containing 4 |
| chr6_+_23712911 | 1.15 |
ENSDART00000167795
|
zgc:158654
|
zgc:158654 |
| chr16_-_39477746 | 1.14 |
ENSDART00000102525
|
stt3b
|
STT3B, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr17_-_2721336 | 1.14 |
ENSDART00000109582
ENSDART00000192691 ENSDART00000189381 |
spata7
|
spermatogenesis associated 7 |
| chr22_-_6219674 | 1.13 |
ENSDART00000182582
|
si:ch211-274k16.2
|
si:ch211-274k16.2 |
| chr10_+_8968203 | 1.13 |
ENSDART00000110443
ENSDART00000080772 |
fstb
|
follistatin b |
| chr10_-_43294933 | 1.12 |
ENSDART00000062631
|
hapln1b
|
hyaluronan and proteoglycan link protein 1b |
| chr20_+_23408970 | 1.12 |
ENSDART00000144261
|
fryl
|
furry homolog, like |
| chr7_+_31074636 | 1.10 |
ENSDART00000173805
|
tjp1a
|
tight junction protein 1a |
| chr23_+_41200854 | 1.10 |
ENSDART00000109567
|
nhsa
|
Nance-Horan syndrome a (congenital cataracts and dental anomalies) |
| chr25_+_25464630 | 1.09 |
ENSDART00000150537
|
rassf7a
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7a |
| chr18_-_16181952 | 1.08 |
ENSDART00000157824
|
slc6a15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr10_-_39283883 | 1.06 |
ENSDART00000023831
|
cry5
|
cryptochrome circadian clock 5 |
| chr23_+_44580254 | 1.05 |
ENSDART00000185069
|
pfn1
|
profilin 1 |
| chr17_-_33289304 | 1.05 |
ENSDART00000135118
ENSDART00000040346 |
efr3ba
|
EFR3 homolog Ba (S. cerevisiae) |
| chr24_-_40860603 | 1.04 |
ENSDART00000188032
|
CU633479.7
|
|
| chr23_+_42434348 | 1.03 |
ENSDART00000161027
|
cyp2aa1
|
cytochrome P450, family 2, subfamily AA, polypeptide 1 |
| chr25_+_3788443 | 1.03 |
ENSDART00000189747
|
chid1
|
chitinase domain containing 1 |
| chr15_-_40267485 | 1.02 |
ENSDART00000152253
|
kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr9_-_28990649 | 1.02 |
ENSDART00000078823
|
ptpn4a
|
protein tyrosine phosphatase, non-receptor type 4a |
| chr23_+_22785375 | 1.01 |
ENSDART00000142085
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr7_+_36552725 | 1.01 |
ENSDART00000173682
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr8_+_26268100 | 1.01 |
ENSDART00000138882
|
slc26a6
|
solute carrier family 26, member 6 |
| chr20_+_26394324 | 1.00 |
ENSDART00000078093
|
zbtb2b
|
zinc finger and BTB domain containing 2b |
| chr19_-_25519612 | 1.00 |
ENSDART00000133150
|
C1GALT1 (1 of many)
|
si:dkey-202e17.1 |
| chr19_-_6873107 | 1.00 |
ENSDART00000124440
|
CABZ01029822.1
|
|
| chr15_+_3125136 | 1.00 |
ENSDART00000130968
|
rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr2_+_33052169 | 1.00 |
ENSDART00000180008
|
rnf220a
|
ring finger protein 220a |
| chr10_-_43540196 | 0.99 |
ENSDART00000170891
|
si:ch73-215f7.1
|
si:ch73-215f7.1 |
| chr5_-_42180205 | 0.99 |
ENSDART00000145247
|
fam222ba
|
family with sequence similarity 222, member Ba |
| chr1_-_51474974 | 0.98 |
ENSDART00000152719
|
spred2a
|
sprouty-related, EVH1 domain containing 2a |
| chr18_+_17959681 | 0.98 |
ENSDART00000142700
|
znf423
|
zinc finger protein 423 |
| chr4_-_5795309 | 0.97 |
ENSDART00000039987
|
pgm3
|
phosphoglucomutase 3 |
| chr13_+_30506781 | 0.96 |
ENSDART00000110884
|
zmiz1a
|
zinc finger, MIZ-type containing 1a |
| chr21_+_26733529 | 0.96 |
ENSDART00000168379
|
pcxa
|
pyruvate carboxylase a |
| chr16_-_30655980 | 0.96 |
ENSDART00000146508
|
ldlrad4b
|
low density lipoprotein receptor class A domain containing 4b |
| chr12_+_5530247 | 0.95 |
ENSDART00000114637
|
ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| chr16_-_8280885 | 0.95 |
ENSDART00000129068
|
entpd3
|
ectonucleoside triphosphate diphosphohydrolase 3 |
| chr20_-_42100932 | 0.95 |
ENSDART00000191930
|
slc35f1
|
solute carrier family 35, member F1 |
| chr10_+_10210455 | 0.94 |
ENSDART00000144214
|
sh2d3ca
|
SH2 domain containing 3Ca |
Network of associatons between targets according to the STRING database.
First level regulatory network of znf740b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.0 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 1.0 | 4.0 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) Wnt signaling pathway involved in somitogenesis(GO:0090244) negative regulation of canonical Wnt signaling pathway involved in heart development(GO:1905067) |
| 0.7 | 2.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.7 | 2.0 | GO:0090278 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.7 | 2.7 | GO:0051610 | serotonin transport(GO:0006837) serotonin uptake(GO:0051610) |
| 0.6 | 4.9 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.6 | 2.3 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.5 | 2.6 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.5 | 1.9 | GO:0015677 | copper ion import(GO:0015677) |
| 0.5 | 1.8 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.4 | 2.5 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.4 | 1.1 | GO:0060031 | mediolateral intercalation(GO:0060031) |
| 0.4 | 2.5 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.3 | 2.4 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.3 | 3.2 | GO:0031272 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.3 | 3.5 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.3 | 4.2 | GO:0051963 | regulation of synapse assembly(GO:0051963) |
| 0.3 | 1.8 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.3 | 6.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.3 | 1.1 | GO:0061033 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.3 | 1.1 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.3 | 1.9 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.3 | 1.9 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.3 | 1.0 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.2 | 1.5 | GO:1902915 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.2 | 3.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 1.8 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.2 | 1.4 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.2 | 1.6 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.2 | 8.2 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 4.4 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.2 | 3.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.2 | 0.9 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) |
| 0.2 | 1.0 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.2 | 2.3 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.2 | 3.0 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.2 | 1.4 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.1 | 6.9 | GO:0045103 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.1 | 2.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 4.3 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 1.0 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 1.0 | GO:0090311 | regulation of histone deacetylation(GO:0031063) regulation of protein deacetylation(GO:0090311) |
| 0.1 | 1.7 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 1.3 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 1.8 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.0 | GO:0046958 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.1 | 3.0 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.1 | 0.6 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.1 | 1.3 | GO:0042664 | locus ceruleus development(GO:0021703) negative regulation of endodermal cell fate specification(GO:0042664) |
| 0.1 | 2.1 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 3.5 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 1.3 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.1 | 2.4 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 1.9 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) post-translational protein modification(GO:0043687) |
| 0.1 | 1.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 0.7 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 1.1 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.9 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.9 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.1 | 0.4 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.1 | 0.4 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.1 | 0.3 | GO:0003242 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 1.9 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 2.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 0.4 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.1 | 1.1 | GO:0097499 | protein localization to nonmotile primary cilium(GO:0097499) |
| 0.1 | 3.9 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 1.0 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 1.7 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 3.4 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.1 | 0.6 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 3.8 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 1.0 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.4 | GO:0032261 | purine nucleotide salvage(GO:0032261) |
| 0.1 | 0.4 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 1.4 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 1.3 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.1 | 0.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.9 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.1 | 0.4 | GO:2001286 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 3.4 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 1.1 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.5 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.5 | GO:0071451 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 1.2 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.9 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 1.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.5 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 1.1 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.6 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 1.3 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 1.3 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.3 | GO:0046247 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 3.6 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 1.9 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 0.3 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.0 | 0.2 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 0.3 | GO:0050890 | cognition(GO:0050890) |
| 0.0 | 0.6 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 1.0 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 1.2 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 3.4 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 0.3 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.0 | 1.7 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 5.7 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 2.1 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.2 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.9 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.7 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 1.8 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 2.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.5 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 1.7 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 2.3 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.7 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 4.4 | GO:0048701 | embryonic cranial skeleton morphogenesis(GO:0048701) |
| 0.0 | 1.7 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 3.4 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 1.0 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.5 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 1.6 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 1.9 | GO:0060027 | convergent extension involved in gastrulation(GO:0060027) |
| 0.0 | 0.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.5 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.6 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.4 | GO:0019319 | hexose biosynthetic process(GO:0019319) |
| 0.0 | 2.3 | GO:0050770 | regulation of axonogenesis(GO:0050770) |
| 0.0 | 0.3 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.4 | GO:0039020 | pronephric nephron tubule development(GO:0039020) |
| 0.0 | 2.1 | GO:0060828 | regulation of canonical Wnt signaling pathway(GO:0060828) |
| 0.0 | 0.4 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 1.6 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.2 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.9 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.3 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 1.2 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 0.8 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 1.1 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.4 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.6 | 4.0 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.5 | 3.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.5 | 1.5 | GO:0032997 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.5 | 3.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.5 | 4.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.3 | 3.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.3 | 2.5 | GO:0035060 | brahma complex(GO:0035060) |
| 0.3 | 1.2 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 3.5 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 1.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.9 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 3.8 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 2.5 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.1 | 2.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.4 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.1 | 9.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 2.8 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.1 | 2.1 | GO:0005686 | U2 snRNP(GO:0005686) U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 1.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 1.9 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 0.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 4.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 3.0 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 2.3 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 1.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 4.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 4.6 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 3.5 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 10.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.9 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 1.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.1 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 1.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.4 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 0.9 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 1.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 1.1 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 1.6 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 3.4 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 6.8 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.9 | 2.7 | GO:0005335 | serotonin:sodium symporter activity(GO:0005335) serotonin transmembrane transporter activity(GO:0015222) |
| 0.6 | 4.9 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.5 | 2.0 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.5 | 1.5 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) IgE receptor activity(GO:0019767) |
| 0.5 | 2.5 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.5 | 1.9 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.5 | 1.9 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.4 | 1.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.4 | 2.6 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.4 | 1.1 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.3 | 1.0 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.3 | 1.2 | GO:0005461 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.3 | 0.9 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.3 | 3.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 1.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.2 | 1.9 | GO:0005113 | patched binding(GO:0005113) |
| 0.2 | 1.0 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 2.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 2.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.2 | 2.4 | GO:0008061 | chitin binding(GO:0008061) |
| 0.2 | 3.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.2 | 2.3 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.2 | 1.8 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 2.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 3.0 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.8 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 1.6 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 4.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 3.0 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 2.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 2.9 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 1.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 1.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 6.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 0.6 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 3.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 7.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.9 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 4.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 1.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 1.8 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.7 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.1 | 0.7 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 1.8 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 1.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.0 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 4.3 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 0.4 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.1 | 3.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 1.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 3.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 3.9 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 1.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 1.0 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 0.2 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.1 | 0.5 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 1.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.9 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 2.0 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.1 | 1.2 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 1.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 1.7 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.9 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 10.2 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.5 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 1.7 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.2 | GO:0047192 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.4 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.5 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 1.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.3 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.3 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 1.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 1.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 5.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.4 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.0 | 1.0 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 4.4 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 2.9 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 0.3 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 0.2 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 1.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.0 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 6.3 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 1.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.2 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 1.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 7.5 | GO:0003779 | actin binding(GO:0003779) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.0 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.2 | 2.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 2.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 1.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 2.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 3.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 2.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 0.6 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 1.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 2.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.1 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.6 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.3 | 2.1 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.3 | 4.0 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.2 | 1.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 4.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 2.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 3.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 3.5 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 2.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.8 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 0.6 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.3 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.1 | 1.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.7 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 2.1 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 2.1 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 1.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |