Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
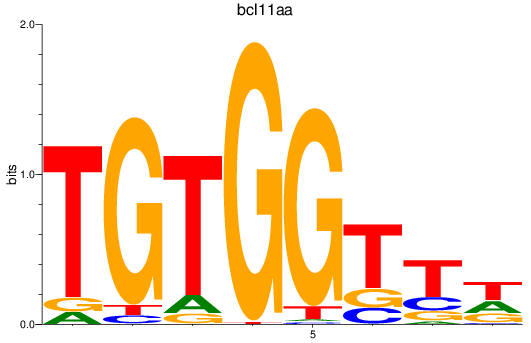
Results for bcl11aa
Z-value: 0.32
Transcription factors associated with bcl11aa
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
bcl11aa
|
ENSDARG00000061352 | BAF chromatin remodeling complex subunit BCL11A a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| bcl11aa | dr11_v1_chr13_+_25853757_25853757 | 0.18 | 6.5e-01 | Click! |
Activity profile of bcl11aa motif
Sorted Z-values of bcl11aa motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_38786298 | 0.39 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr7_-_34265481 | 0.38 |
ENSDART00000173596
|
si:ch211-98n17.5
|
si:ch211-98n17.5 |
| chr2_+_4207209 | 0.36 |
ENSDART00000157903
ENSDART00000166476 |
gata6
|
GATA binding protein 6 |
| chr3_-_19367081 | 0.35 |
ENSDART00000191369
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr24_-_21921262 | 0.35 |
ENSDART00000186061
ENSDART00000187846 |
tagln3b
|
transgelin 3b |
| chr9_-_14504834 | 0.32 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr4_+_12031958 | 0.32 |
ENSDART00000044154
|
tnnt2c
|
troponin T2c, cardiac |
| chr8_+_47188154 | 0.31 |
ENSDART00000137319
|
si:dkeyp-100a1.6
|
si:dkeyp-100a1.6 |
| chr6_-_7438584 | 0.28 |
ENSDART00000053776
|
fkbp11
|
FK506 binding protein 11 |
| chr17_+_15433518 | 0.28 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr22_+_21255860 | 0.27 |
ENSDART00000134893
|
cpamd8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8 |
| chr17_+_15433671 | 0.27 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr2_+_48288461 | 0.27 |
ENSDART00000141495
|
hes6
|
hes family bHLH transcription factor 6 |
| chr6_+_30668098 | 0.26 |
ENSDART00000112294
|
ttc22
|
tetratricopeptide repeat domain 22 |
| chr3_+_28576173 | 0.26 |
ENSDART00000151189
|
sept12
|
septin 12 |
| chr13_-_6081803 | 0.26 |
ENSDART00000099224
|
dld
|
deltaD |
| chr19_+_30633453 | 0.26 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr11_+_8129536 | 0.25 |
ENSDART00000158112
ENSDART00000011183 |
prkacba
|
protein kinase, cAMP-dependent, catalytic, beta a |
| chr18_+_40354998 | 0.25 |
ENSDART00000098791
ENSDART00000049171 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr15_-_26636826 | 0.25 |
ENSDART00000087632
|
slc47a4
|
solute carrier family 47 (multidrug and toxin extrusion), member 4 |
| chr6_+_45918981 | 0.24 |
ENSDART00000149642
|
h6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr16_-_29527412 | 0.24 |
ENSDART00000175376
|
onecutl
|
one cut domain, family member, like |
| chr11_+_30057762 | 0.24 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr2_+_36898982 | 0.23 |
ENSDART00000084859
|
rabgap1l2
|
RAB GTPase activating protein 1-like 2 |
| chr5_-_28606916 | 0.22 |
ENSDART00000026107
ENSDART00000137717 |
tnc
|
tenascin C |
| chr6_+_34868156 | 0.22 |
ENSDART00000149364
|
il23r
|
interleukin 23 receptor |
| chr8_+_44703864 | 0.21 |
ENSDART00000016225
|
star
|
steroidogenic acute regulatory protein |
| chr15_-_34567370 | 0.19 |
ENSDART00000099793
|
sostdc1a
|
sclerostin domain containing 1a |
| chr25_+_14017609 | 0.19 |
ENSDART00000129105
ENSDART00000125733 |
chst1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 |
| chr7_-_2163361 | 0.19 |
ENSDART00000173654
|
si:cabz01007812.1
|
si:cabz01007812.1 |
| chr21_+_5915041 | 0.19 |
ENSDART00000151370
|
prodha
|
proline dehydrogenase (oxidase) 1a |
| chr7_-_29571615 | 0.19 |
ENSDART00000019140
|
rorab
|
RAR-related orphan receptor A, paralog b |
| chr10_-_43771447 | 0.18 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr17_-_26868169 | 0.18 |
ENSDART00000157204
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr5_+_34622320 | 0.18 |
ENSDART00000141338
|
enc1
|
ectodermal-neural cortex 1 |
| chr20_+_52554352 | 0.18 |
ENSDART00000153217
ENSDART00000145230 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr10_+_38526496 | 0.17 |
ENSDART00000144329
|
acer3
|
alkaline ceramidase 3 |
| chr5_-_31901468 | 0.17 |
ENSDART00000147814
ENSDART00000141446 |
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr8_-_31369161 | 0.16 |
ENSDART00000019937
|
gadd45ga
|
growth arrest and DNA-damage-inducible, gamma a |
| chr11_+_4026229 | 0.15 |
ENSDART00000041417
|
camk1b
|
calcium/calmodulin-dependent protein kinase Ib |
| chr13_+_24834199 | 0.15 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr4_+_16710001 | 0.15 |
ENSDART00000035899
|
pkp2
|
plakophilin 2 |
| chr16_+_8716800 | 0.14 |
ENSDART00000124693
ENSDART00000181961 |
cabz01093075.1
|
cabz01093075.1 |
| chr13_-_47403154 | 0.14 |
ENSDART00000114318
|
bcl2l11
|
BCL2 like 11 |
| chr11_-_29685853 | 0.13 |
ENSDART00000128468
|
nr0b1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr19_+_14059349 | 0.13 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr18_-_24988645 | 0.13 |
ENSDART00000136434
ENSDART00000085735 |
chd2
|
chromodomain helicase DNA binding protein 2 |
| chr11_+_38280454 | 0.13 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr20_+_46216431 | 0.13 |
ENSDART00000185384
|
stx7l
|
syntaxin 7-like |
| chr17_+_26569601 | 0.13 |
ENSDART00000153897
|
ndnfl
|
neuron-derived neurotrophic factor , like |
| chr22_-_20309283 | 0.12 |
ENSDART00000182125
ENSDART00000048775 |
si:dkey-110c1.10
|
si:dkey-110c1.10 |
| chr25_-_14637660 | 0.12 |
ENSDART00000143666
|
nav2b
|
neuron navigator 2b |
| chr14_+_22022441 | 0.12 |
ENSDART00000149121
|
clcf1
|
cardiotrophin-like cytokine factor 1 |
| chr25_+_18954189 | 0.12 |
ENSDART00000123207
|
tdg.2
|
thymine DNA glycosylase, tandem duplicate 2 |
| chr20_+_15015557 | 0.12 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr2_+_32796873 | 0.12 |
ENSDART00000077511
|
ccr9a
|
chemokine (C-C motif) receptor 9a |
| chr12_+_28856151 | 0.12 |
ENSDART00000152969
|
nfe2l1b
|
nuclear factor, erythroid 2-like 1b |
| chr11_+_30058139 | 0.12 |
ENSDART00000112254
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr4_+_34126849 | 0.12 |
ENSDART00000162442
|
si:ch211-223g7.6
|
si:ch211-223g7.6 |
| chr19_+_29798064 | 0.12 |
ENSDART00000167803
ENSDART00000051804 |
marcksl1b
|
MARCKS-like 1b |
| chr20_+_34543365 | 0.11 |
ENSDART00000152073
|
si:ch211-242b18.1
|
si:ch211-242b18.1 |
| chr24_+_31361407 | 0.11 |
ENSDART00000162668
|
cremb
|
cAMP responsive element modulator b |
| chr18_+_40525408 | 0.11 |
ENSDART00000132833
|
vwa7
|
von Willebrand factor A domain containing 7 |
| chr5_-_38122126 | 0.11 |
ENSDART00000141791
ENSDART00000170528 |
si:ch211-284e13.6
|
si:ch211-284e13.6 |
| chr2_-_5505094 | 0.11 |
ENSDART00000145035
|
saga
|
S-antigen; retina and pineal gland (arrestin) a |
| chr9_+_13986427 | 0.11 |
ENSDART00000147200
|
cd28
|
CD28 molecule |
| chr19_-_22478888 | 0.10 |
ENSDART00000090679
|
pleca
|
plectin a |
| chr23_+_40908583 | 0.10 |
ENSDART00000180933
|
LO017845.1
|
|
| chr17_-_44776224 | 0.10 |
ENSDART00000156195
|
zdhhc22
|
zinc finger, DHHC-type containing 22 |
| chr4_-_23858900 | 0.10 |
ENSDART00000123199
|
usp6nl
|
USP6 N-terminal like |
| chr2_-_31302615 | 0.10 |
ENSDART00000034784
ENSDART00000060812 |
adcyap1b
|
adenylate cyclase activating polypeptide 1b |
| chr11_+_23957440 | 0.09 |
ENSDART00000190721
|
cntn2
|
contactin 2 |
| chr12_-_19279103 | 0.09 |
ENSDART00000186669
|
si:ch211-141o9.10
|
si:ch211-141o9.10 |
| chr19_+_40350468 | 0.09 |
ENSDART00000087444
|
hepacam2
|
HEPACAM family member 2 |
| chr3_-_56512473 | 0.09 |
ENSDART00000125439
ENSDART00000181743 |
cyth1a
|
cytohesin 1a |
| chr24_-_36680261 | 0.09 |
ENSDART00000059507
|
ccr10
|
chemokine (C-C motif) receptor 10 |
| chr18_+_35742838 | 0.09 |
ENSDART00000088504
ENSDART00000140386 |
rasgrp4
|
RAS guanyl releasing protein 4 |
| chr18_-_2639351 | 0.09 |
ENSDART00000168106
|
relt
|
RELT, TNF receptor |
| chr2_+_24786765 | 0.09 |
ENSDART00000141030
|
pde4ca
|
phosphodiesterase 4C, cAMP-specific a |
| chr7_-_7984015 | 0.08 |
ENSDART00000163203
|
si:cabz01030277.1
|
si:cabz01030277.1 |
| chr20_-_14054083 | 0.08 |
ENSDART00000009549
|
rhag
|
Rh associated glycoprotein |
| chr4_+_5215950 | 0.08 |
ENSDART00000140613
|
galnt8b.2
|
polypeptide N-acetylgalactosaminyltransferase 8b, tandem duplicate 2 |
| chr15_-_36352983 | 0.08 |
ENSDART00000059786
|
si:dkey-23k10.5
|
si:dkey-23k10.5 |
| chr4_-_5595237 | 0.08 |
ENSDART00000109854
|
vegfab
|
vascular endothelial growth factor Ab |
| chr24_+_11334733 | 0.08 |
ENSDART00000147552
ENSDART00000143171 |
si:dkey-12l12.1
|
si:dkey-12l12.1 |
| chr6_-_18960105 | 0.08 |
ENSDART00000185278
ENSDART00000162968 |
sept9b
|
septin 9b |
| chr16_+_30483043 | 0.07 |
ENSDART00000188034
|
pear1
|
platelet endothelial aggregation receptor 1 |
| chr19_-_22488952 | 0.07 |
ENSDART00000179856
ENSDART00000141503 |
pleca
|
plectin a |
| chr16_+_19014886 | 0.07 |
ENSDART00000079298
|
si:ch211-254p10.2
|
si:ch211-254p10.2 |
| chr20_+_40150612 | 0.07 |
ENSDART00000143680
ENSDART00000109681 ENSDART00000101041 ENSDART00000121818 |
trdn
|
triadin |
| chr1_-_41982582 | 0.07 |
ENSDART00000014678
|
adra1d
|
adrenoceptor alpha 1D |
| chr9_+_41612642 | 0.07 |
ENSDART00000138473
|
spegb
|
SPEG complex locus b |
| chr17_+_19730001 | 0.07 |
ENSDART00000155025
|
grem2a
|
gremlin 2, DAN family BMP antagonist a |
| chr7_+_49865049 | 0.07 |
ENSDART00000164165
|
slc1a2a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2a |
| chr22_-_20379045 | 0.07 |
ENSDART00000183511
|
zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr2_+_22694382 | 0.06 |
ENSDART00000139196
|
kif1ab
|
kinesin family member 1Ab |
| chr9_-_44939104 | 0.06 |
ENSDART00000192903
|
vil1
|
villin 1 |
| chr4_+_20255160 | 0.06 |
ENSDART00000188658
|
lrtm2a
|
leucine-rich repeats and transmembrane domains 2a |
| chr17_+_23770848 | 0.06 |
ENSDART00000079646
|
kcnk18
|
potassium channel, subfamily K, member 18 |
| chr19_+_19786117 | 0.06 |
ENSDART00000167757
ENSDART00000163546 |
hoxa1a
|
homeobox A1a |
| chr16_+_10963602 | 0.06 |
ENSDART00000141032
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr13_+_31108334 | 0.06 |
ENSDART00000142245
|
arhgap22
|
Rho GTPase activating protein 22 |
| chr4_+_3980247 | 0.06 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr1_+_33217911 | 0.05 |
ENSDART00000136761
|
prkx
|
protein kinase, X-linked |
| chr16_+_11724230 | 0.05 |
ENSDART00000060266
|
ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr17_+_15882533 | 0.05 |
ENSDART00000164124
|
gabrr2a
|
gamma-aminobutyric acid (GABA) A receptor, rho 2a |
| chr1_+_23398405 | 0.05 |
ENSDART00000102646
|
rhoh
|
ras homolog family member H |
| chr7_+_46368520 | 0.05 |
ENSDART00000192821
|
znf536
|
zinc finger protein 536 |
| chr6_-_19023468 | 0.05 |
ENSDART00000184729
|
sept9b
|
septin 9b |
| chr24_+_13635108 | 0.05 |
ENSDART00000183008
|
trpa1b
|
transient receptor potential cation channel, subfamily A, member 1b |
| chr19_-_7144548 | 0.05 |
ENSDART00000147177
ENSDART00000134850 |
psmb8a
psmb13a
|
proteasome subunit beta 8A proteasome subunit beta 13a |
| chr3_+_30968176 | 0.05 |
ENSDART00000186266
|
prf1.9
|
perforin 1.9 |
| chr6_+_53288018 | 0.05 |
ENSDART00000170319
|
cysltr3
|
cysteinyl leukotriene receptor 3 |
| chr4_-_4261673 | 0.04 |
ENSDART00000150694
|
cd9b
|
CD9 molecule b |
| chr1_+_23406403 | 0.04 |
ENSDART00000189921
|
chrna9
|
cholinergic receptor, nicotinic, alpha 9 |
| chr14_-_30967284 | 0.04 |
ENSDART00000149435
|
il2rgb
|
interleukin 2 receptor, gamma b |
| chr11_+_7580079 | 0.04 |
ENSDART00000091550
ENSDART00000193223 ENSDART00000193386 |
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr17_+_5426087 | 0.04 |
ENSDART00000131430
ENSDART00000188636 |
runx2a
|
runt-related transcription factor 2a |
| chr21_-_21096437 | 0.04 |
ENSDART00000186552
|
ank1b
|
ankyrin 1, erythrocytic b |
| chr8_+_25342896 | 0.04 |
ENSDART00000129032
|
CR847543.1
|
|
| chr17_+_5351922 | 0.04 |
ENSDART00000105394
|
runx2a
|
runt-related transcription factor 2a |
| chr2_-_24369087 | 0.04 |
ENSDART00000081237
|
plvapa
|
plasmalemma vesicle associated protein a |
| chr1_+_56873359 | 0.04 |
ENSDART00000152713
|
si:ch211-152f2.2
|
si:ch211-152f2.2 |
| chr9_-_22352280 | 0.04 |
ENSDART00000115384
ENSDART00000060361 |
crygm5
|
crystallin, gamma M5 |
| chr10_-_15128771 | 0.04 |
ENSDART00000101261
|
spp1
|
secreted phosphoprotein 1 |
| chr5_+_13359146 | 0.04 |
ENSDART00000179839
|
ccl19a.1
|
chemokine (C-C motif) ligand 19a, tandem duplicate 1 |
| chr9_-_31108285 | 0.04 |
ENSDART00000003193
|
gpr183a
|
G protein-coupled receptor 183a |
| chr25_-_35996141 | 0.04 |
ENSDART00000149074
|
sall1b
|
spalt-like transcription factor 1b |
| chr17_+_8899016 | 0.03 |
ENSDART00000150214
|
psmc1a
|
proteasome 26S subunit, ATPase 1a |
| chr15_-_38154616 | 0.03 |
ENSDART00000099392
|
irgq2
|
immunity-related GTPase family, q2 |
| chr21_+_27283054 | 0.03 |
ENSDART00000139058
|
si:dkey-175m17.7
|
si:dkey-175m17.7 |
| chr10_-_23358357 | 0.03 |
ENSDART00000135475
|
cadm2a
|
cell adhesion molecule 2a |
| chr20_-_36809059 | 0.03 |
ENSDART00000062925
|
slc25a27
|
solute carrier family 25, member 27 |
| chr10_+_41199660 | 0.03 |
ENSDART00000125314
|
adrb3b
|
adrenoceptor beta 3b |
| chr7_+_39706004 | 0.03 |
ENSDART00000161856
|
ccl36.1
|
chemokine (C-C motif) ligand 36, duplicate 1 |
| chr17_-_8899323 | 0.03 |
ENSDART00000081590
|
nkl.1
|
NK-lysin tandem duplicate 1 |
| chr25_+_18953756 | 0.03 |
ENSDART00000154291
|
tdg.2
|
thymine DNA glycosylase, tandem duplicate 2 |
| chr25_-_22889519 | 0.03 |
ENSDART00000128250
|
mob2a
|
MOB kinase activator 2a |
| chr3_-_21382491 | 0.03 |
ENSDART00000016562
|
itga2b
|
integrin, alpha 2b |
| chr19_+_3849378 | 0.03 |
ENSDART00000166218
ENSDART00000159228 |
oscp1a
|
organic solute carrier partner 1a |
| chr11_+_24703108 | 0.03 |
ENSDART00000159173
|
gpr25
|
G protein-coupled receptor 25 |
| chr25_+_34845469 | 0.03 |
ENSDART00000145416
|
tmem231
|
transmembrane protein 231 |
| chr8_+_36803415 | 0.02 |
ENSDART00000111680
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr22_-_15578402 | 0.02 |
ENSDART00000062986
|
hsh2d
|
hematopoietic SH2 domain containing |
| chr22_-_5323482 | 0.02 |
ENSDART00000145785
|
s1pr4
|
sphingosine-1-phosphate receptor 4 |
| chr10_-_28395620 | 0.02 |
ENSDART00000168907
|
CR392341.4
|
|
| chr16_-_17300030 | 0.02 |
ENSDART00000149267
|
kel
|
Kell blood group, metallo-endopeptidase |
| chr24_+_13635387 | 0.02 |
ENSDART00000105997
|
trpa1b
|
transient receptor potential cation channel, subfamily A, member 1b |
| chr7_-_17337233 | 0.02 |
ENSDART00000050236
ENSDART00000102141 |
nitr8
|
novel immune-type receptor 8 |
| chr14_-_6727717 | 0.02 |
ENSDART00000166979
|
si:dkeyp-44a8.4
|
si:dkeyp-44a8.4 |
| chr21_-_11632403 | 0.02 |
ENSDART00000171708
ENSDART00000138619 ENSDART00000136308 ENSDART00000144770 |
cast
|
calpastatin |
| chr2_-_36925561 | 0.02 |
ENSDART00000187690
|
map1sb
|
microtubule-associated protein 1Sb |
| chr5_+_24156170 | 0.02 |
ENSDART00000136570
|
slc25a15b
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15b |
| chr11_+_25139495 | 0.01 |
ENSDART00000168368
|
si:ch211-25d12.7
|
si:ch211-25d12.7 |
| chr3_+_30968883 | 0.01 |
ENSDART00000085736
|
prf1.9
|
perforin 1.9 |
| chr9_+_30112423 | 0.01 |
ENSDART00000112398
ENSDART00000013591 |
tfg
|
trk-fused gene |
| chr24_-_31306724 | 0.01 |
ENSDART00000165399
|
acp5b
|
acid phosphatase 5b, tartrate resistant |
| chr25_-_3470910 | 0.01 |
ENSDART00000029067
ENSDART00000186737 |
hbp1
|
HMG-box transcription factor 1 |
| chr7_-_17258635 | 0.01 |
ENSDART00000173934
ENSDART00000173668 ENSDART00000161797 |
nitr12
|
novel immune-type receptor 12 |
| chr5_-_41860556 | 0.01 |
ENSDART00000140154
|
si:dkey-65b12.12
|
si:dkey-65b12.12 |
| chr3_+_51660158 | 0.01 |
ENSDART00000155512
ENSDART00000171764 |
chmp6a
|
charged multivesicular body protein 6a |
| chr3_-_45298487 | 0.01 |
ENSDART00000102245
|
pdpk1a
|
3-phosphoinositide dependent protein kinase 1a |
| chr7_-_30553588 | 0.01 |
ENSDART00000139546
|
sltm
|
SAFB-like, transcription modulator |
| chr16_-_22863348 | 0.01 |
ENSDART00000147609
|
shc1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr24_+_37722048 | 0.01 |
ENSDART00000191918
|
rab11fip3
|
RAB11 family interacting protein 3 (class II) |
| chr16_+_27957808 | 0.01 |
ENSDART00000133696
|
znf804b
|
zinc finger protein 804B |
| chr9_+_9296222 | 0.01 |
ENSDART00000141325
|
si:ch211-214p13.8
|
si:ch211-214p13.8 |
| chr1_+_57741577 | 0.01 |
ENSDART00000192673
|
LO018597.1
|
|
| chr5_+_28313824 | 0.01 |
ENSDART00000135878
|
si:dkeyp-86b9.1
|
si:dkeyp-86b9.1 |
| chr8_-_43997538 | 0.00 |
ENSDART00000186449
|
rimbp2
|
RIMS binding protein 2 |
| chr1_+_11736420 | 0.00 |
ENSDART00000140329
|
chst12b
|
carbohydrate (chondroitin 4) sulfotransferase 12b |
| chr25_-_27564590 | 0.00 |
ENSDART00000073508
|
hyal4
|
hyaluronoglucosaminidase 4 |
| chr16_-_22863603 | 0.00 |
ENSDART00000098400
ENSDART00000183963 |
shc1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr3_+_54761569 | 0.00 |
ENSDART00000135913
ENSDART00000180983 |
si:ch211-74m13.1
|
si:ch211-74m13.1 |
| chr7_+_17534485 | 0.00 |
ENSDART00000170911
|
nitr1k
|
novel immune-type receptor 1k |
| chr21_+_11923701 | 0.00 |
ENSDART00000109292
|
ubap2a
|
ubiquitin associated protein 2a |
| chr1_+_39597809 | 0.00 |
ENSDART00000122700
|
tenm3
|
teneurin transmembrane protein 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of bcl11aa
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.1 | 0.4 | GO:0001773 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.3 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.2 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.1 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 0.1 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.0 | 0.1 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.3 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.0 | 0.1 | GO:0007635 | chemosensory behavior(GO:0007635) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.1 | GO:0008584 | male gonad development(GO:0008584) |
| 0.0 | 0.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.2 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:2000392 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.0 | 0.2 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.1 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.0 | 0.1 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.2 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.1 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.0 | 0.0 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.3 | GO:0010737 | protein kinase A signaling(GO:0010737) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043218 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.1 | GO:0014704 | intercalated disc(GO:0014704) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.4 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.3 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.3 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 0.0 | 0.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0008263 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.0 | 0.2 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.0 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |