Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
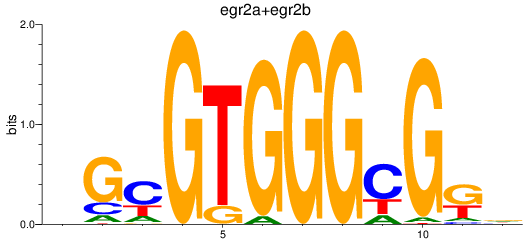
Results for egr2a+egr2b
Z-value: 0.67
Transcription factors associated with egr2a+egr2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
egr2b
|
ENSDARG00000042826 | early growth response 2b |
|
egr2a
|
ENSDARG00000044098 | early growth response 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| egr2b | dr11_v1_chr12_-_8504278_8504278 | 0.06 | 8.7e-01 | Click! |
| egr2a | dr11_v1_chr17_-_43666166_43666166 | -0.05 | 8.9e-01 | Click! |
Activity profile of egr2a+egr2b motif
Sorted Z-values of egr2a+egr2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_29556844 | 0.94 |
ENSDART00000138021
|
rbp7a
|
retinol binding protein 7a, cellular |
| chr8_-_53490376 | 0.81 |
ENSDART00000158789
|
chdh
|
choline dehydrogenase |
| chr6_-_39313027 | 0.74 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr3_+_32492467 | 0.74 |
ENSDART00000151329
|
trpm4a
|
transient receptor potential cation channel, subfamily M, member 4a |
| chr11_+_1796426 | 0.72 |
ENSDART00000173330
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr6_-_15604417 | 0.64 |
ENSDART00000157817
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr14_+_7939398 | 0.55 |
ENSDART00000189773
|
cxxc5b
|
CXXC finger protein 5b |
| chr1_-_7659870 | 0.53 |
ENSDART00000085203
|
efnb2b
|
ephrin-B2b |
| chr3_-_39488482 | 0.53 |
ENSDART00000135192
|
zgc:100868
|
zgc:100868 |
| chr3_-_6767440 | 0.52 |
ENSDART00000156174
|
mast1b
|
microtubule associated serine/threonine kinase 1b |
| chr7_-_58776400 | 0.51 |
ENSDART00000167433
|
sox17
|
SRY (sex determining region Y)-box 17 |
| chr23_+_20422661 | 0.51 |
ENSDART00000144047
ENSDART00000104336 |
tnnc2
|
troponin C type 2 (fast) |
| chr23_+_25856541 | 0.50 |
ENSDART00000145426
ENSDART00000028236 |
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr17_-_52643970 | 0.50 |
ENSDART00000190594
|
spred1
|
sprouty-related, EVH1 domain containing 1 |
| chr3_-_39488639 | 0.48 |
ENSDART00000161644
|
zgc:100868
|
zgc:100868 |
| chr8_-_51753604 | 0.48 |
ENSDART00000007090
|
tbx16
|
T-box 16 |
| chr25_-_29134654 | 0.48 |
ENSDART00000067066
|
parp6b
|
poly (ADP-ribose) polymerase family, member 6b |
| chr5_+_51026563 | 0.47 |
ENSDART00000050988
|
gcnt4a
|
glucosaminyl (N-acetyl) transferase 4, core 2, a |
| chr12_-_30777540 | 0.47 |
ENSDART00000126466
|
entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr15_-_16012963 | 0.46 |
ENSDART00000144138
|
hnf1ba
|
HNF1 homeobox Ba |
| chr10_+_10351685 | 0.46 |
ENSDART00000109432
|
cercam
|
cerebral endothelial cell adhesion molecule |
| chr16_+_40575742 | 0.45 |
ENSDART00000161503
|
ccne2
|
cyclin E2 |
| chr6_+_56147812 | 0.44 |
ENSDART00000150219
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr12_+_11080776 | 0.44 |
ENSDART00000079336
|
raraa
|
retinoic acid receptor, alpha a |
| chr12_+_695619 | 0.42 |
ENSDART00000161691
|
abcc3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr3_+_12322170 | 0.41 |
ENSDART00000161227
|
glis2b
|
GLIS family zinc finger 2b |
| chr23_+_37815645 | 0.41 |
ENSDART00000011627
|
irx7
|
iroquois homeobox 7 |
| chr12_-_30777743 | 0.41 |
ENSDART00000148888
|
entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr7_-_29534001 | 0.41 |
ENSDART00000124028
|
anxa2b
|
annexin A2b |
| chr23_-_39849155 | 0.41 |
ENSDART00000115330
|
ppp1r14c
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr5_-_57655092 | 0.40 |
ENSDART00000074290
|
mia
|
melanoma inhibitory activity |
| chr14_+_36220479 | 0.39 |
ENSDART00000148319
|
pitx2
|
paired-like homeodomain 2 |
| chr7_-_33023404 | 0.38 |
ENSDART00000052383
|
cd81a
|
CD81 molecule a |
| chr16_-_26676685 | 0.38 |
ENSDART00000103431
|
esrp1
|
epithelial splicing regulatory protein 1 |
| chr1_-_411331 | 0.38 |
ENSDART00000092524
|
rasa3
|
RAS p21 protein activator 3 |
| chr16_-_6821927 | 0.38 |
ENSDART00000149070
ENSDART00000149570 |
mbpb
|
myelin basic protein b |
| chr2_+_33326522 | 0.37 |
ENSDART00000056655
|
klf17
|
Kruppel-like factor 17 |
| chr11_-_7139555 | 0.36 |
ENSDART00000016472
|
bmp7a
|
bone morphogenetic protein 7a |
| chr10_+_38643304 | 0.36 |
ENSDART00000067447
|
mmp30
|
matrix metallopeptidase 30 |
| chr14_+_909769 | 0.36 |
ENSDART00000021346
ENSDART00000172777 |
arl3l2
|
ADP-ribosylation factor-like 3, like 2 |
| chr21_+_53504 | 0.36 |
ENSDART00000170452
|
dmgdh
|
dimethylglycine dehydrogenase |
| chr16_+_23978978 | 0.34 |
ENSDART00000058964
ENSDART00000135084 |
apoa2
|
apolipoprotein A-II |
| chr2_+_26288301 | 0.34 |
ENSDART00000017668
|
ptbp1a
|
polypyrimidine tract binding protein 1a |
| chr11_+_24251141 | 0.34 |
ENSDART00000182684
|
pnp4a
|
purine nucleoside phosphorylase 4a |
| chr8_-_52859301 | 0.33 |
ENSDART00000162004
|
nr5a1a
|
nuclear receptor subfamily 5, group A, member 1a |
| chr9_-_9671244 | 0.33 |
ENSDART00000018228
|
gsk3b
|
glycogen synthase kinase 3 beta |
| chr24_+_35564668 | 0.32 |
ENSDART00000122734
|
cebpd
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr25_-_18125769 | 0.32 |
ENSDART00000140484
|
kitlga
|
kit ligand a |
| chr7_-_5125799 | 0.32 |
ENSDART00000173390
|
ltb4r2a
|
leukotriene B4 receptor 2a |
| chr5_+_36513605 | 0.31 |
ENSDART00000013590
|
wnt11
|
wingless-type MMTV integration site family, member 11 |
| chr20_+_37844035 | 0.31 |
ENSDART00000041397
|
flvcr1
|
feline leukemia virus subgroup C cellular receptor 1 |
| chr16_+_13424534 | 0.30 |
ENSDART00000114944
|
zgc:101640
|
zgc:101640 |
| chr5_+_22969651 | 0.30 |
ENSDART00000089992
ENSDART00000145477 |
hmgn7
|
high mobility group nucleosomal binding domain 7 |
| chr5_-_40510397 | 0.30 |
ENSDART00000146237
ENSDART00000051065 |
fsta
|
follistatin a |
| chr10_-_15053507 | 0.30 |
ENSDART00000157446
ENSDART00000170441 |
CLDN23
|
si:ch211-95j8.5 |
| chr4_+_30775376 | 0.30 |
ENSDART00000158528
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr19_-_31341850 | 0.29 |
ENSDART00000040810
|
arl4ab
|
ADP-ribosylation factor-like 4ab |
| chr20_+_13894123 | 0.29 |
ENSDART00000007744
|
slc30a1a
|
solute carrier family 30 (zinc transporter), member 1a |
| chr15_+_46344655 | 0.29 |
ENSDART00000155893
|
si:ch1073-340i21.2
|
si:ch1073-340i21.2 |
| chr20_-_53366137 | 0.28 |
ENSDART00000146001
|
wasf1
|
WAS protein family, member 1 |
| chr5_+_32791245 | 0.28 |
ENSDART00000077189
|
ier5l
|
immediate early response 5-like |
| chr6_-_15604157 | 0.28 |
ENSDART00000141597
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr16_+_51180938 | 0.28 |
ENSDART00000169022
|
hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr19_+_43119698 | 0.28 |
ENSDART00000167847
ENSDART00000186962 ENSDART00000187305 |
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr9_-_33121535 | 0.28 |
ENSDART00000166371
ENSDART00000138052 |
zgc:172014
|
zgc:172014 |
| chr7_-_26408472 | 0.28 |
ENSDART00000111494
|
gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr2_-_22535 | 0.28 |
ENSDART00000157877
|
CABZ01092282.1
|
|
| chr23_+_23232136 | 0.27 |
ENSDART00000126479
ENSDART00000187764 |
plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr18_-_3166726 | 0.27 |
ENSDART00000165002
|
aqp11
|
aquaporin 11 |
| chr17_+_27456804 | 0.26 |
ENSDART00000017756
ENSDART00000181461 ENSDART00000180178 |
ctsl.1
|
cathepsin L.1 |
| chr23_+_39606108 | 0.26 |
ENSDART00000109464
|
g0s2
|
G0/G1 switch 2 |
| chr23_-_637347 | 0.26 |
ENSDART00000132175
|
l1camb
|
L1 cell adhesion molecule, paralog b |
| chr16_+_4658250 | 0.26 |
ENSDART00000006212
|
LIN28A
|
si:ch1073-284b18.2 |
| chr20_-_26420939 | 0.25 |
ENSDART00000110883
|
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr14_+_29769336 | 0.25 |
ENSDART00000105898
|
si:dkey-34l15.1
|
si:dkey-34l15.1 |
| chr19_-_617246 | 0.25 |
ENSDART00000062551
|
cyp51
|
cytochrome P450, family 51 |
| chr13_+_25428677 | 0.25 |
ENSDART00000186284
|
si:dkey-51a16.9
|
si:dkey-51a16.9 |
| chr18_+_22109379 | 0.25 |
ENSDART00000147230
|
zgc:158868
|
zgc:158868 |
| chr20_-_26421112 | 0.25 |
ENSDART00000183767
ENSDART00000182330 |
akap12b
|
A kinase (PRKA) anchor protein 12b |
| chr6_-_35446110 | 0.25 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr4_+_1600034 | 0.25 |
ENSDART00000146779
|
slc38a2
|
solute carrier family 38, member 2 |
| chr16_-_12512568 | 0.24 |
ENSDART00000055161
ENSDART00000160906 |
cox6b2
|
cytochrome c oxidase subunit VIb polypeptide 2 |
| chr23_-_2081554 | 0.24 |
ENSDART00000179805
|
ndnf
|
neuron-derived neurotrophic factor |
| chr19_+_1964005 | 0.24 |
ENSDART00000172049
|
sh3bp5a
|
SH3-domain binding protein 5a (BTK-associated) |
| chr15_+_42397125 | 0.23 |
ENSDART00000169751
|
tiam1b
|
T cell lymphoma invasion and metastasis 1b |
| chr9_+_17348745 | 0.23 |
ENSDART00000147488
|
slain1a
|
SLAIN motif family, member 1a |
| chr8_+_25267903 | 0.23 |
ENSDART00000093090
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr20_+_52554352 | 0.23 |
ENSDART00000153217
ENSDART00000145230 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr23_+_9560797 | 0.23 |
ENSDART00000180014
|
adrm1
|
adhesion regulating molecule 1 |
| chr16_+_28754403 | 0.23 |
ENSDART00000103340
|
s100v1
|
S100 calcium binding protein V1 |
| chr8_+_34731982 | 0.23 |
ENSDART00000066050
|
hpdb
|
4-hydroxyphenylpyruvate dioxygenase b |
| chr6_-_16406210 | 0.22 |
ENSDART00000012023
|
faimb
|
Fas apoptotic inhibitory molecule b |
| chr5_+_68807170 | 0.22 |
ENSDART00000017849
|
her7
|
hairy and enhancer of split related-7 |
| chr11_+_3254524 | 0.22 |
ENSDART00000159459
|
pmela
|
premelanosome protein a |
| chr19_+_10493223 | 0.22 |
ENSDART00000180826
|
si:ch211-171h4.6
|
si:ch211-171h4.6 |
| chr20_-_9760424 | 0.22 |
ENSDART00000104936
|
si:dkey-63j12.4
|
si:dkey-63j12.4 |
| chr23_+_9560991 | 0.22 |
ENSDART00000081433
ENSDART00000131594 ENSDART00000130069 ENSDART00000138601 |
adrm1
|
adhesion regulating molecule 1 |
| chr18_+_30028135 | 0.22 |
ENSDART00000140825
ENSDART00000145306 ENSDART00000136810 |
si:ch211-220f16.1
|
si:ch211-220f16.1 |
| chr11_-_33919309 | 0.22 |
ENSDART00000078202
|
phka2
|
phosphorylase kinase, alpha 2 (liver) |
| chr7_-_10560964 | 0.22 |
ENSDART00000172761
ENSDART00000170476 |
mthfs
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chr20_-_45661049 | 0.21 |
ENSDART00000124582
ENSDART00000131251 |
napbb
|
N-ethylmaleimide-sensitive factor attachment protein, beta b |
| chr23_-_2513300 | 0.21 |
ENSDART00000067652
|
snai1b
|
snail family zinc finger 1b |
| chr13_+_12739283 | 0.21 |
ENSDART00000102279
|
lingo2b
|
leucine rich repeat and Ig domain containing 2b |
| chr4_-_9173552 | 0.21 |
ENSDART00000042963
|
chst11
|
carbohydrate (chondroitin 4) sulfotransferase 11 |
| chr13_-_15865335 | 0.21 |
ENSDART00000135186
|
si:ch211-57f7.7
|
si:ch211-57f7.7 |
| chr15_+_1397811 | 0.21 |
ENSDART00000102125
|
schip1
|
schwannomin interacting protein 1 |
| chr12_+_23762966 | 0.21 |
ENSDART00000152942
ENSDART00000181725 |
jcada
|
junctional cadherin 5 associated a |
| chr1_+_55293424 | 0.21 |
ENSDART00000152464
|
zgc:172106
|
zgc:172106 |
| chr25_-_33275666 | 0.20 |
ENSDART00000193605
|
CABZ01046954.1
|
|
| chr25_-_23526058 | 0.20 |
ENSDART00000191331
ENSDART00000062930 |
phlda2
|
pleckstrin homology-like domain, family A, member 2 |
| chr9_+_3035128 | 0.20 |
ENSDART00000140693
|
si:ch211-173m16.2
|
si:ch211-173m16.2 |
| chr23_+_42338325 | 0.20 |
ENSDART00000169660
|
cyp2aa7
|
cytochrome P450, family 2, subfamily AA, polypeptide 7 |
| chr2_-_17828279 | 0.20 |
ENSDART00000083314
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr25_+_1304173 | 0.20 |
ENSDART00000155229
|
rxfp3.3b
|
relaxin/insulin-like family peptide receptor 3.3b |
| chr21_+_28958471 | 0.20 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr14_+_31865324 | 0.20 |
ENSDART00000039880
|
tm9sf5
|
transmembrane 9 superfamily protein member 5 |
| chr4_+_16710001 | 0.19 |
ENSDART00000035899
|
pkp2
|
plakophilin 2 |
| chr15_+_34699585 | 0.19 |
ENSDART00000017569
|
tnfb
|
tumor necrosis factor b (TNF superfamily, member 2) |
| chr2_-_17827983 | 0.19 |
ENSDART00000166518
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr8_+_53051701 | 0.19 |
ENSDART00000131514
|
nadka
|
NAD kinase a |
| chr17_+_2162916 | 0.19 |
ENSDART00000103775
|
pak6a
|
p21 protein (Cdc42/Rac)-activated kinase 6a |
| chr9_-_21231297 | 0.19 |
ENSDART00000162578
|
pla1a
|
phospholipase A1 member A |
| chr11_+_3254252 | 0.18 |
ENSDART00000123568
|
pmela
|
premelanosome protein a |
| chr18_-_39702327 | 0.18 |
ENSDART00000149158
|
dmxl2
|
Dmx-like 2 |
| chr23_+_43489182 | 0.18 |
ENSDART00000162062
|
znf341
|
zinc finger protein 341 |
| chr19_+_636886 | 0.18 |
ENSDART00000149192
|
tert
|
telomerase reverse transcriptase |
| chr25_+_34013093 | 0.18 |
ENSDART00000011967
|
anxa2a
|
annexin A2a |
| chr21_+_30950097 | 0.18 |
ENSDART00000187572
|
rhogb
|
ras homolog family member Gb |
| chr1_-_20271138 | 0.18 |
ENSDART00000185931
|
ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr8_+_49975160 | 0.18 |
ENSDART00000156403
ENSDART00000080135 |
gfpt1
|
glutamine--fructose-6-phosphate transaminase 1 |
| chr12_-_49168398 | 0.18 |
ENSDART00000186608
|
FO704624.1
|
|
| chr6_+_42818963 | 0.18 |
ENSDART00000184833
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr16_+_11029762 | 0.18 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr17_+_53435279 | 0.18 |
ENSDART00000126630
|
cx52.9
|
connexin 52.9 |
| chr7_+_25053331 | 0.18 |
ENSDART00000173998
|
si:dkey-23i12.7
|
si:dkey-23i12.7 |
| chr11_-_24191928 | 0.18 |
ENSDART00000136827
|
sox12
|
SRY (sex determining region Y)-box 12 |
| chr6_+_42819337 | 0.17 |
ENSDART00000046498
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr5_-_26093945 | 0.17 |
ENSDART00000010199
ENSDART00000145096 |
fam219ab
|
family with sequence similarity 219, member Ab |
| chr1_-_8653385 | 0.17 |
ENSDART00000193041
|
actb1
|
actin, beta 1 |
| chr9_-_30274412 | 0.17 |
ENSDART00000089526
|
otc
|
ornithine carbamoyltransferase |
| chr17_+_12698532 | 0.17 |
ENSDART00000064509
ENSDART00000136830 |
stmn4l
|
stathmin-like 4, like |
| chr21_-_7882905 | 0.17 |
ENSDART00000056561
|
s100z
|
S100 calcium binding protein Z |
| chr5_+_46293850 | 0.17 |
ENSDART00000140636
|
si:ch211-130m23.2
|
si:ch211-130m23.2 |
| chr18_+_402048 | 0.17 |
ENSDART00000166345
|
gpib
|
glucose-6-phosphate isomerase b |
| chr16_+_68069 | 0.16 |
ENSDART00000185385
ENSDART00000159652 |
sox4b
|
SRY (sex determining region Y)-box 4b |
| chr4_+_63484571 | 0.16 |
ENSDART00000169518
ENSDART00000168681 |
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr13_-_39947335 | 0.16 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr3_-_61185746 | 0.16 |
ENSDART00000028219
|
pvalb4
|
parvalbumin 4 |
| chr24_+_2519761 | 0.16 |
ENSDART00000106619
|
nrn1a
|
neuritin 1a |
| chr10_+_15777064 | 0.16 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr4_-_58846245 | 0.16 |
ENSDART00000170777
|
si:dkey-28i19.1
|
si:dkey-28i19.1 |
| chr13_+_31144305 | 0.15 |
ENSDART00000189602
|
CR931802.4
|
|
| chr22_+_38581012 | 0.15 |
ENSDART00000169239
|
BX088728.1
|
|
| chr20_+_13969414 | 0.15 |
ENSDART00000049864
|
rd3
|
retinal degeneration 3 |
| chr14_-_36863432 | 0.15 |
ENSDART00000158052
|
rnf130
|
ring finger protein 130 |
| chr22_-_6651382 | 0.15 |
ENSDART00000124308
|
si:ch211-209l18.4
|
si:ch211-209l18.4 |
| chr3_+_23737795 | 0.15 |
ENSDART00000182247
|
hoxb3a
|
homeobox B3a |
| chr18_+_41495841 | 0.15 |
ENSDART00000098671
|
si:ch211-203b8.6
|
si:ch211-203b8.6 |
| chr11_+_37251825 | 0.15 |
ENSDART00000169804
|
il17rc
|
interleukin 17 receptor C |
| chr5_-_68022631 | 0.15 |
ENSDART00000143199
|
wasf3a
|
WAS protein family, member 3a |
| chr11_-_236766 | 0.15 |
ENSDART00000163978
|
dusp7
|
dual specificity phosphatase 7 |
| chr21_+_11415224 | 0.15 |
ENSDART00000049036
|
zgc:92275
|
zgc:92275 |
| chr24_+_3963684 | 0.15 |
ENSDART00000182959
ENSDART00000185926 ENSDART00000167043 ENSDART00000033394 |
pfkpa
|
phosphofructokinase, platelet a |
| chr3_-_38918697 | 0.15 |
ENSDART00000145630
|
si:dkey-106c17.2
|
si:dkey-106c17.2 |
| chr3_+_23692462 | 0.15 |
ENSDART00000145934
|
hoxb7a
|
homeobox B7a |
| chr11_-_236984 | 0.15 |
ENSDART00000170778
|
dusp7
|
dual specificity phosphatase 7 |
| chr17_+_53311618 | 0.15 |
ENSDART00000166517
|
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr5_-_72136548 | 0.15 |
ENSDART00000007827
|
spra
|
sepiapterin reductase a |
| chr16_-_12173399 | 0.14 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr1_-_22861348 | 0.14 |
ENSDART00000139412
|
SMIM18
|
si:dkey-92j12.6 |
| chr24_-_31439841 | 0.14 |
ENSDART00000169952
|
cngb3.2
|
cyclic nucleotide gated channel beta 3, tandem duplicate 2 |
| chr3_-_30685401 | 0.14 |
ENSDART00000151097
|
si:ch211-51c14.1
|
si:ch211-51c14.1 |
| chr2_-_7431590 | 0.14 |
ENSDART00000185699
|
asip2b
|
agouti signaling protein, nonagouti homolog (mouse) 2b |
| chr10_+_252425 | 0.14 |
ENSDART00000059478
|
lrrc32
|
leucine rich repeat containing 32 |
| chr12_+_13282797 | 0.14 |
ENSDART00000137757
ENSDART00000152397 |
irf9
|
interferon regulatory factor 9 |
| chr7_-_69636502 | 0.14 |
ENSDART00000126739
|
tspan5a
|
tetraspanin 5a |
| chr3_+_40315410 | 0.14 |
ENSDART00000083241
ENSDART00000132827 |
slc29a4
|
solute carrier family 29 (equilibrative nucleoside transporter), member 4 |
| chr11_+_7324704 | 0.13 |
ENSDART00000031937
|
diras1a
|
DIRAS family, GTP-binding RAS-like 1a |
| chr22_-_18746508 | 0.13 |
ENSDART00000003929
|
midn
|
midnolin |
| chr3_+_62217479 | 0.13 |
ENSDART00000175012
|
BX470259.4
|
|
| chr2_-_37874647 | 0.13 |
ENSDART00000039386
|
zgc:66427
|
zgc:66427 |
| chr1_+_47459723 | 0.13 |
ENSDART00000015046
|
fstl1a
|
follistatin-like 1a |
| chr1_-_59176949 | 0.13 |
ENSDART00000128742
|
CABZ01118678.1
|
|
| chr5_+_43870389 | 0.13 |
ENSDART00000141002
|
zgc:112966
|
zgc:112966 |
| chr8_+_6576940 | 0.13 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr11_+_329687 | 0.13 |
ENSDART00000172882
|
cyp27b1
|
cytochrome P450, family 27, subfamily B, polypeptide 1 |
| chr15_-_22074315 | 0.13 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr5_-_37252111 | 0.13 |
ENSDART00000185110
|
lrch2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr13_+_51853716 | 0.13 |
ENSDART00000175341
ENSDART00000187855 |
LT631684.1
|
|
| chr25_+_37348730 | 0.13 |
ENSDART00000156639
|
pamr1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr2_+_30147504 | 0.13 |
ENSDART00000190947
|
kcnb2
|
potassium voltage-gated channel, Shab-related subfamily, member 2 |
| chr2_-_155270 | 0.13 |
ENSDART00000131177
|
adcy1b
|
adenylate cyclase 1b |
| chr1_+_44941031 | 0.13 |
ENSDART00000141145
|
si:dkey-9i23.16
|
si:dkey-9i23.16 |
| chr1_+_57187794 | 0.13 |
ENSDART00000152485
|
si:dkey-27j5.9
|
si:dkey-27j5.9 |
| chr14_+_918287 | 0.12 |
ENSDART00000167066
|
CU462913.1
|
|
| chr3_-_46818001 | 0.12 |
ENSDART00000166505
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr3_+_60007703 | 0.12 |
ENSDART00000157351
ENSDART00000153928 ENSDART00000155876 |
si:ch211-110e21.3
|
si:ch211-110e21.3 |
| chr15_+_31526225 | 0.12 |
ENSDART00000154456
|
wdr95
|
WD40 repeat domain 95 |
| chr15_-_1687204 | 0.12 |
ENSDART00000155579
ENSDART00000181382 |
sptssb
|
serine palmitoyltransferase, small subunit B |
| chr19_-_10196370 | 0.12 |
ENSDART00000091707
|
dbpa
|
D site albumin promoter binding protein a |
Network of associatons between targets according to the STRING database.
First level regulatory network of egr2a+egr2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.2 | 0.7 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.2 | 0.5 | GO:0032060 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.2 | 0.5 | GO:0048341 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) intermediate mesoderm development(GO:0048389) |
| 0.1 | 0.9 | GO:0033986 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.1 | 0.4 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 0.4 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.1 | 0.3 | GO:0010312 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.1 | 0.3 | GO:0060471 | negative regulation of fertilization(GO:0060467) prevention of polyspermy(GO:0060468) cortical granule exocytosis(GO:0060471) |
| 0.1 | 0.5 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 0.4 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.1 | 0.4 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.1 | 0.5 | GO:0021571 | rhombomere 5 development(GO:0021571) rhombomere 6 development(GO:0021572) |
| 0.1 | 0.3 | GO:0097037 | heme export(GO:0097037) |
| 0.1 | 0.3 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 0.2 | GO:1901546 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.1 | 0.2 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 0.2 | GO:0021856 | pallium development(GO:0021543) cerebral cortex cell migration(GO:0021795) cerebral cortex tangential migration(GO:0021800) cerebral cortex tangential migration using cell-axon interactions(GO:0021824) substrate-dependent cerebral cortex tangential migration(GO:0021825) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) cerebral cortex development(GO:0021987) |
| 0.1 | 0.2 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.1 | 0.2 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 0.2 | GO:1903589 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.0 | 0.1 | GO:0032369 | negative regulation of lipid transport(GO:0032369) |
| 0.0 | 0.4 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.4 | GO:0021703 | locus ceruleus development(GO:0021703) |
| 0.0 | 0.4 | GO:0071632 | optomotor response(GO:0071632) |
| 0.0 | 0.4 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.2 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.2 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.0 | 0.4 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.2 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.0 | 0.4 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.0 | 0.1 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.2 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.2 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.0 | 0.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.4 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.5 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 0.2 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.3 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0061551 | trigeminal ganglion development(GO:0061551) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.9 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.5 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.2 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.4 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.0 | 0.2 | GO:0061099 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.3 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.0 | 0.3 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.2 | GO:1902042 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 0.1 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.5 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.5 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0001774 | microglial cell activation(GO:0001774) |
| 0.0 | 0.1 | GO:0042559 | tetrahydrobiopterin biosynthetic process(GO:0006729) pteridine-containing compound biosynthetic process(GO:0042559) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.1 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.0 | 0.5 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.0 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.1 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.1 | GO:0030825 | regulation of cGMP metabolic process(GO:0030823) positive regulation of cGMP metabolic process(GO:0030825) regulation of cGMP biosynthetic process(GO:0030826) positive regulation of cGMP biosynthetic process(GO:0030828) regulation of guanylate cyclase activity(GO:0031282) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.0 | 0.5 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.3 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.4 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.2 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.0 | GO:0045023 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.1 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 0.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.2 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.1 | 0.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.2 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.2 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.0 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.1 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.1 | GO:0070062 | extracellular exosome(GO:0070062) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 0.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.4 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.1 | 0.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.2 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.1 | 0.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.2 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.1 | 1.0 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.2 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.6 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 0.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.3 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.2 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.3 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.2 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.2 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.0 | 0.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.3 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.2 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.7 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.0 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.2 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.3 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.1 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.3 | GO:0016248 | ion channel inhibitor activity(GO:0008200) channel inhibitor activity(GO:0016248) |
| 0.0 | 0.1 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.7 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.3 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |