Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
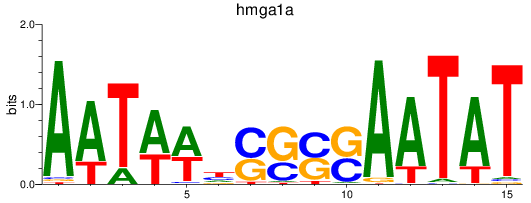
Results for hmga1a
Z-value: 0.68
Transcription factors associated with hmga1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hmga1a
|
ENSDARG00000028335 | high mobility group AT-hook 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hmga1a | dr11_v1_chr23_-_3759692_3759692 | 0.74 | 2.4e-02 | Click! |
Activity profile of hmga1a motif
Sorted Z-values of hmga1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_20195350 | 0.73 |
ENSDART00000139675
|
dao.1
|
D-amino-acid oxidase, tandem duplicate 1 |
| chr25_-_34670413 | 0.68 |
ENSDART00000073440
|
DNAJA4
|
DnaJ heat shock protein family (Hsp40) member A4 |
| chr12_-_656540 | 0.66 |
ENSDART00000172651
|
sult2st2
|
sulfotransferase family 2, cytosolic sulfotransferase 2 |
| chr3_+_62161184 | 0.61 |
ENSDART00000090370
ENSDART00000192665 |
noxo1a
|
NADPH oxidase organizer 1a |
| chr23_-_45439903 | 0.59 |
ENSDART00000170729
|
NPNT (1 of many)
|
nephronectin |
| chr16_+_1383914 | 0.59 |
ENSDART00000185089
|
cers2b
|
ceramide synthase 2b |
| chr8_-_50981175 | 0.54 |
ENSDART00000004065
|
zgc:91909
|
zgc:91909 |
| chr23_+_25354856 | 0.53 |
ENSDART00000109023
ENSDART00000147440 |
fmnl3
|
formin-like 3 |
| chr15_-_29387446 | 0.50 |
ENSDART00000145976
ENSDART00000035096 |
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr12_-_10409961 | 0.50 |
ENSDART00000149521
ENSDART00000052001 |
eef2k
|
eukaryotic elongation factor 2 kinase |
| chr7_+_33130639 | 0.46 |
ENSDART00000142450
ENSDART00000173967 ENSDART00000173832 |
zgc:153219
si:ch211-194p6.7
|
zgc:153219 si:ch211-194p6.7 |
| chr16_-_29557338 | 0.45 |
ENSDART00000058888
|
hormad1
|
HORMA domain containing 1 |
| chr6_-_58764672 | 0.44 |
ENSDART00000154322
|
soat2
|
sterol O-acyltransferase 2 |
| chr23_-_28347039 | 0.43 |
ENSDART00000145072
|
neurod4
|
neuronal differentiation 4 |
| chr5_-_38451082 | 0.40 |
ENSDART00000136428
|
chrne
|
cholinergic receptor, nicotinic, epsilon |
| chr21_+_22845317 | 0.38 |
ENSDART00000065555
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr2_+_25839193 | 0.38 |
ENSDART00000078634
|
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr20_+_34400715 | 0.36 |
ENSDART00000061632
|
fam129aa
|
family with sequence similarity 129, member Aa |
| chr7_-_22956716 | 0.34 |
ENSDART00000122113
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr7_-_22956889 | 0.34 |
ENSDART00000101447
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr17_-_40110782 | 0.32 |
ENSDART00000126929
|
si:dkey-187k19.2
|
si:dkey-187k19.2 |
| chr11_-_16975190 | 0.30 |
ENSDART00000122222
|
suclg2
|
succinate-CoA ligase, GDP-forming, beta subunit |
| chr5_+_32831561 | 0.29 |
ENSDART00000169358
ENSDART00000192078 |
si:ch211-208h16.4
|
si:ch211-208h16.4 |
| chr16_+_2433696 | 0.29 |
ENSDART00000182803
|
CU928063.1
|
|
| chr18_+_14595546 | 0.28 |
ENSDART00000080788
|
wfdc1
|
WAP four-disulfide core domain 1 |
| chr6_-_58757131 | 0.28 |
ENSDART00000083582
|
soat2
|
sterol O-acyltransferase 2 |
| chr15_+_47440477 | 0.27 |
ENSDART00000002384
|
phox2a
|
paired-like homeobox 2a |
| chr23_-_45407631 | 0.25 |
ENSDART00000148484
ENSDART00000150186 |
zgc:101853
|
zgc:101853 |
| chr9_+_25568839 | 0.24 |
ENSDART00000177342
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr7_+_15871156 | 0.24 |
ENSDART00000145946
|
pax6b
|
paired box 6b |
| chr17_+_51520073 | 0.23 |
ENSDART00000189646
ENSDART00000170951 ENSDART00000189492 |
pxdn
|
peroxidasin |
| chr13_+_49175947 | 0.23 |
ENSDART00000056927
|
egln1a
|
egl-9 family hypoxia-inducible factor 1a |
| chr12_-_36740781 | 0.23 |
ENSDART00000105484
|
si:ch211-216b21.2
|
si:ch211-216b21.2 |
| chr25_+_581227 | 0.23 |
ENSDART00000126863
|
nell2a
|
neural EGFL like 2a |
| chr7_-_16558665 | 0.22 |
ENSDART00000169399
|
csrp3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr11_+_37095165 | 0.22 |
ENSDART00000124997
|
card19
|
caspase recruitment domain family, member 19 |
| chr13_+_49175624 | 0.21 |
ENSDART00000193550
|
egln1a
|
egl-9 family hypoxia-inducible factor 1a |
| chr23_+_45512825 | 0.21 |
ENSDART00000064846
|
prelid1b
|
PRELI domain containing 1b |
| chr17_-_5769196 | 0.20 |
ENSDART00000113885
|
si:dkey-100n19.2
|
si:dkey-100n19.2 |
| chr4_+_17245217 | 0.20 |
ENSDART00000184215
ENSDART00000110908 |
casc1
|
cancer susceptibility candidate 1 |
| chr4_+_65537216 | 0.19 |
ENSDART00000181458
|
si:dkey-205i10.1
|
si:dkey-205i10.1 |
| chr14_-_33521071 | 0.19 |
ENSDART00000052789
|
c1galt1c1
|
C1GALT1-specific chaperone 1 |
| chr4_+_17245005 | 0.19 |
ENSDART00000027645
|
casc1
|
cancer susceptibility candidate 1 |
| chr23_-_27633730 | 0.18 |
ENSDART00000103639
|
arf3a
|
ADP-ribosylation factor 3a |
| chr18_+_14595805 | 0.18 |
ENSDART00000167825
|
wfdc1
|
WAP four-disulfide core domain 1 |
| chr10_+_42733210 | 0.17 |
ENSDART00000189832
|
CABZ01063556.1
|
|
| chr24_-_35534273 | 0.17 |
ENSDART00000026578
|
ube2v2
|
ubiquitin-conjugating enzyme E2 variant 2 |
| chr21_-_20765338 | 0.17 |
ENSDART00000135940
|
ghrb
|
growth hormone receptor b |
| chr13_+_1089942 | 0.17 |
ENSDART00000054322
|
cnrip1b
|
cannabinoid receptor interacting protein 1b |
| chr6_+_22679610 | 0.16 |
ENSDART00000102701
|
zmp:0000000634
|
zmp:0000000634 |
| chr13_+_41917606 | 0.16 |
ENSDART00000114741
|
polr1b
|
polymerase (RNA) I polypeptide B |
| chr17_-_21200406 | 0.16 |
ENSDART00000104708
|
abhd12
|
abhydrolase domain containing 12 |
| chr14_-_38827442 | 0.15 |
ENSDART00000160000
|
spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr19_+_29808471 | 0.15 |
ENSDART00000186428
|
hdac1
|
histone deacetylase 1 |
| chr8_+_49975160 | 0.15 |
ENSDART00000156403
ENSDART00000080135 |
gfpt1
|
glutamine--fructose-6-phosphate transaminase 1 |
| chr11_-_4235811 | 0.14 |
ENSDART00000121716
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr16_-_12236362 | 0.14 |
ENSDART00000114759
|
lpcat3
|
lysophosphatidylcholine acyltransferase 3 |
| chr18_+_328689 | 0.14 |
ENSDART00000167841
|
ss18l2
|
synovial sarcoma translocation gene on chromosome 18-like 2 |
| chr8_+_40644838 | 0.14 |
ENSDART00000169311
|
adra2b
|
adrenoceptor alpha 2B |
| chr19_+_29808699 | 0.13 |
ENSDART00000051799
ENSDART00000164205 |
hdac1
|
histone deacetylase 1 |
| chr20_+_26349002 | 0.13 |
ENSDART00000152842
|
syne1a
|
spectrin repeat containing, nuclear envelope 1a |
| chr5_-_20814576 | 0.13 |
ENSDART00000098682
ENSDART00000147639 |
si:ch211-225b11.1
|
si:ch211-225b11.1 |
| chr24_-_29963858 | 0.13 |
ENSDART00000183442
|
CR352310.1
|
|
| chr15_+_888704 | 0.13 |
ENSDART00000182796
|
si:dkey-7i4.9
|
si:dkey-7i4.9 |
| chr24_-_18477672 | 0.13 |
ENSDART00000126928
ENSDART00000158393 |
si:dkey-73n8.3
|
si:dkey-73n8.3 |
| chr16_-_42969598 | 0.13 |
ENSDART00000156011
|
si:ch211-135n15.3
|
si:ch211-135n15.3 |
| chr3_+_26019426 | 0.12 |
ENSDART00000135389
ENSDART00000182411 |
foxred2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr3_+_36515376 | 0.12 |
ENSDART00000161652
|
si:dkeyp-72e1.9
|
si:dkeyp-72e1.9 |
| chr4_-_992063 | 0.12 |
ENSDART00000181630
ENSDART00000183898 ENSDART00000160902 |
naga
|
N-acetylgalactosaminidase, alpha |
| chr7_+_48999723 | 0.11 |
ENSDART00000182699
ENSDART00000166329 |
si:ch211-288d18.1
|
si:ch211-288d18.1 |
| chr24_-_31306724 | 0.10 |
ENSDART00000165399
|
acp5b
|
acid phosphatase 5b, tartrate resistant |
| chr11_-_12051502 | 0.10 |
ENSDART00000183462
|
socs7
|
suppressor of cytokine signaling 7 |
| chr11_-_12051283 | 0.09 |
ENSDART00000170516
|
socs7
|
suppressor of cytokine signaling 7 |
| chr6_-_2222707 | 0.09 |
ENSDART00000022179
|
prkag1
|
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr6_-_7123210 | 0.08 |
ENSDART00000041304
|
atg3
|
autophagy related 3 |
| chr10_-_1726148 | 0.08 |
ENSDART00000187527
|
gal3st1b
|
galactose-3-O-sulfotransferase 1b |
| chr14_-_38828057 | 0.08 |
ENSDART00000186088
|
spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr5_+_20421539 | 0.08 |
ENSDART00000164499
|
selplg
|
selectin P ligand |
| chr4_+_72668095 | 0.07 |
ENSDART00000182282
ENSDART00000162637 |
CABZ01054391.1
|
|
| chr4_-_45028479 | 0.07 |
ENSDART00000143150
ENSDART00000076870 |
si:dkey-51d8.1
|
si:dkey-51d8.1 |
| chr10_-_1725699 | 0.07 |
ENSDART00000183555
|
gal3st1b
|
galactose-3-O-sulfotransferase 1b |
| chr21_+_7545062 | 0.07 |
ENSDART00000023997
|
tbca
|
tubulin cofactor a |
| chr8_-_42579446 | 0.06 |
ENSDART00000189792
|
dok2
|
docking protein 2 |
| chr12_-_44196222 | 0.06 |
ENSDART00000171730
|
si:ch73-329n5.1
|
si:ch73-329n5.1 |
| chr14_+_8710568 | 0.06 |
ENSDART00000169505
|
kcnk4a
|
potassium channel, subfamily K, member 4a |
| chr5_-_66296635 | 0.06 |
ENSDART00000125993
|
prlrb
|
prolactin receptor b |
| chr14_+_46020571 | 0.05 |
ENSDART00000157617
ENSDART00000083928 |
c1qtnf2
|
C1q and TNF related 2 |
| chr4_+_13568469 | 0.05 |
ENSDART00000171235
ENSDART00000136152 |
calua
|
calumenin a |
| chr1_+_2112726 | 0.05 |
ENSDART00000131714
ENSDART00000138396 |
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr17_-_15201031 | 0.05 |
ENSDART00000124426
|
socs4
|
suppressor of cytokine signaling 4 |
| chr6_+_8630355 | 0.05 |
ENSDART00000161749
ENSDART00000193976 |
tspeara
|
thrombospondin-type laminin G domain and EAR repeats a |
| chr1_+_44360973 | 0.05 |
ENSDART00000167568
|
si:ch211-165a10.5
|
si:ch211-165a10.5 |
| chr5_-_51830997 | 0.05 |
ENSDART00000163616
|
homer1b
|
homer scaffolding protein 1b |
| chr15_+_17030941 | 0.04 |
ENSDART00000062069
|
plin2
|
perilipin 2 |
| chr15_+_17030473 | 0.04 |
ENSDART00000129407
|
plin2
|
perilipin 2 |
| chr7_-_33130552 | 0.04 |
ENSDART00000127006
|
mns1
|
meiosis-specific nuclear structural 1 |
| chr12_+_46512881 | 0.04 |
ENSDART00000105454
|
CU855711.1
|
|
| chr8_-_53735399 | 0.04 |
ENSDART00000187005
|
tlr9
|
toll-like receptor 9 |
| chr2_-_10062575 | 0.03 |
ENSDART00000091726
|
fam78ba
|
family with sequence similarity 78, member B a |
| chr17_+_31739418 | 0.03 |
ENSDART00000155073
ENSDART00000156180 |
arhgap5
|
Rho GTPase activating protein 5 |
| chr16_+_11652591 | 0.03 |
ENSDART00000133876
|
si:dkey-250k15.9
|
si:dkey-250k15.9 |
| chr16_+_11724230 | 0.03 |
ENSDART00000060266
|
ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr9_-_12885201 | 0.03 |
ENSDART00000124957
|
ankzf1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr25_-_2355107 | 0.02 |
ENSDART00000056121
|
mrps35
|
mitochondrial ribosomal protein S35 |
| chr12_+_46483618 | 0.02 |
ENSDART00000186970
|
BX005305.5
|
|
| chr21_+_40302747 | 0.02 |
ENSDART00000174166
|
si:ch211-218m3.18
|
si:ch211-218m3.18 |
| chr12_+_46425800 | 0.02 |
ENSDART00000191965
|
BX005305.1
|
|
| chr15_+_44135726 | 0.02 |
ENSDART00000192151
|
CU655961.2
|
|
| chr18_-_35407289 | 0.01 |
ENSDART00000012018
|
snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr14_+_46020282 | 0.01 |
ENSDART00000190087
|
c1qtnf2
|
C1q and TNF related 2 |
| chr14_+_34951202 | 0.01 |
ENSDART00000047524
ENSDART00000115105 |
il12ba
|
interleukin 12Ba |
| chr6_+_3827751 | 0.01 |
ENSDART00000003008
ENSDART00000122348 |
gad1b
|
glutamate decarboxylase 1b |
| chr6_-_48094342 | 0.01 |
ENSDART00000137458
|
slc2a1b
|
solute carrier family 2 (facilitated glucose transporter), member 1b |
| chr22_-_10580194 | 0.01 |
ENSDART00000105848
|
SHISA4 (1 of many)
|
si:dkey-42i9.7 |
| chr1_+_19849168 | 0.01 |
ENSDART00000111454
|
si:dkey-82j4.2
|
si:dkey-82j4.2 |
| chr18_-_46380471 | 0.01 |
ENSDART00000144444
ENSDART00000086772 |
slc19a3b
|
solute carrier family 19 (thiamine transporter), member 3b |
| chr14_-_15699528 | 0.00 |
ENSDART00000161123
|
neurl1b
|
neuralized E3 ubiquitin protein ligase 1B |
Network of associatons between targets according to the STRING database.
First level regulatory network of hmga1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0046144 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.1 | 0.5 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.1 | 0.4 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.1 | 0.7 | GO:0034435 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.1 | 0.7 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.1 | 0.4 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.1 | 0.2 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.1 | 0.4 | GO:0045905 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.2 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.3 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.0 | 0.2 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.0 | 0.2 | GO:0003311 | pancreatic D cell differentiation(GO:0003311) pancreatic epsilon cell differentiation(GO:0090104) |
| 0.0 | 0.5 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.2 | GO:0052651 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.2 | GO:0071459 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 0.1 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 0.3 | GO:0021703 | locus ceruleus development(GO:0021703) |
| 0.0 | 0.7 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.6 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.1 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.4 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.1 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 | 0.2 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.1 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.1 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.5 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.2 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.0 | GO:0071706 | tumor necrosis factor production(GO:0032640) tumor necrosis factor superfamily cytokine production(GO:0071706) |
| 0.0 | 0.1 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.2 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.6 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.0 | 0.2 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.2 | 0.7 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.1 | 0.3 | GO:0004775 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.1 | 0.6 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0048531 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.2 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.0 | 0.6 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.4 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.3 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.5 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.4 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |