Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
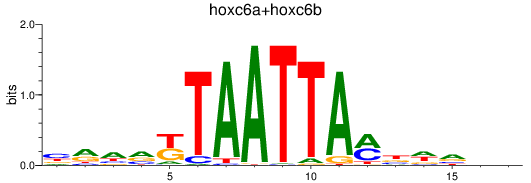
Results for hoxc6a+hoxc6b
Z-value: 1.00
Transcription factors associated with hoxc6a+hoxc6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc6a
|
ENSDARG00000070343 | homeobox C6a |
|
hoxc6b
|
ENSDARG00000101954 | homeobox C6b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc6b | dr11_v1_chr11_+_2198831_2198831 | -0.69 | 4.0e-02 | Click! |
| hoxc6a | dr11_v1_chr23_+_36115541_36115541 | -0.55 | 1.3e-01 | Click! |
Activity profile of hoxc6a+hoxc6b motif
Sorted Z-values of hoxc6a+hoxc6b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_22320738 | 1.76 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr8_-_53044300 | 1.61 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr20_+_54299419 | 1.30 |
ENSDART00000056089
ENSDART00000193107 |
si:zfos-1505d6.3
|
si:zfos-1505d6.3 |
| chr20_+_54309148 | 1.26 |
ENSDART00000099360
|
zp2.1
|
zona pellucida glycoprotein 2, tandem duplicate 1 |
| chr20_+_54304800 | 1.23 |
ENSDART00000121661
|
zp2.6
|
zona pellucida glycoprotein 2, tandem duplicate 6 |
| chr10_-_34916208 | 1.19 |
ENSDART00000187371
|
ccna1
|
cyclin A1 |
| chr25_-_21031007 | 0.99 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr23_+_1029450 | 0.90 |
ENSDART00000189196
|
si:zfos-905g2.1
|
si:zfos-905g2.1 |
| chr2_-_31833347 | 0.87 |
ENSDART00000109460
|
nol7
|
nucleolar protein 7 |
| chr16_+_29509133 | 0.86 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr21_-_44081540 | 0.72 |
ENSDART00000130833
|
FO704810.1
|
|
| chr17_+_30369396 | 0.66 |
ENSDART00000076611
|
greb1
|
growth regulation by estrogen in breast cancer 1 |
| chr25_-_37186894 | 0.66 |
ENSDART00000191647
ENSDART00000182095 |
tdrd12
|
tudor domain containing 12 |
| chr21_+_43172506 | 0.63 |
ENSDART00000121725
|
zcchc10
|
zinc finger, CCHC domain containing 10 |
| chr2_-_15324837 | 0.61 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr7_+_24573721 | 0.61 |
ENSDART00000173938
ENSDART00000173681 |
si:dkeyp-75h12.7
|
si:dkeyp-75h12.7 |
| chr2_+_56657804 | 0.60 |
ENSDART00000113964
|
POLR2E (1 of many)
|
RNA polymerase II subunit E |
| chr2_+_10878406 | 0.58 |
ENSDART00000091497
|
tceanc2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chr22_-_16154771 | 0.57 |
ENSDART00000009464
|
slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr23_-_18024543 | 0.56 |
ENSDART00000139695
|
pm20d1.1
|
peptidase M20 domain containing 1, tandem duplicate 1 |
| chr5_-_37103487 | 0.56 |
ENSDART00000149211
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr15_-_43284021 | 0.55 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr23_-_18707418 | 0.53 |
ENSDART00000144668
ENSDART00000141205 ENSDART00000016765 |
zgc:103759
|
zgc:103759 |
| chr5_+_41477954 | 0.53 |
ENSDART00000185871
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr22_-_36530902 | 0.53 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr14_-_48765262 | 0.53 |
ENSDART00000166463
|
cnot6b
|
CCR4-NOT transcription complex, subunit 6b |
| chr2_+_15048410 | 0.52 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr2_+_31833997 | 0.51 |
ENSDART00000066788
|
epdr1
|
ependymin related 1 |
| chr3_-_19368435 | 0.48 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr13_-_52089003 | 0.48 |
ENSDART00000187600
|
tmem254
|
transmembrane protein 254 |
| chr19_+_10592778 | 0.47 |
ENSDART00000135488
ENSDART00000151624 |
si:dkey-211g8.5
|
si:dkey-211g8.5 |
| chr1_-_9195629 | 0.46 |
ENSDART00000143587
ENSDART00000192174 |
ern2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr7_-_5375214 | 0.46 |
ENSDART00000033316
|
vangl2
|
VANGL planar cell polarity protein 2 |
| chr10_-_7386475 | 0.46 |
ENSDART00000167963
|
nrg1
|
neuregulin 1 |
| chr24_+_34113424 | 0.45 |
ENSDART00000105572
|
gbx1
|
gastrulation brain homeobox 1 |
| chr16_-_7793457 | 0.45 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr24_+_21514283 | 0.45 |
ENSDART00000007066
|
cdk8
|
cyclin-dependent kinase 8 |
| chr3_-_59297532 | 0.44 |
ENSDART00000187991
|
CABZ01053748.1
|
|
| chr4_-_49582758 | 0.44 |
ENSDART00000180834
ENSDART00000187608 |
si:dkey-159n16.2
|
si:dkey-159n16.2 |
| chr6_-_43283122 | 0.43 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr16_-_42965192 | 0.43 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr16_+_54209504 | 0.42 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr11_-_44979281 | 0.42 |
ENSDART00000190972
|
ldb1b
|
LIM-domain binding 1b |
| chr1_+_22851261 | 0.41 |
ENSDART00000193925
|
gtf2e2
|
general transcription factor IIE, polypeptide 2, beta |
| chr8_+_26874924 | 0.41 |
ENSDART00000141794
|
rimkla
|
ribosomal modification protein rimK-like family member A |
| chr24_-_38657683 | 0.40 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr5_+_41477526 | 0.39 |
ENSDART00000153567
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr2_-_10877765 | 0.39 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr5_-_51198430 | 0.39 |
ENSDART00000132503
ENSDART00000097473 ENSDART00000165870 |
snapc4
|
small nuclear RNA activating complex, polypeptide 4 |
| chr17_+_51682429 | 0.38 |
ENSDART00000004379
|
nol10
|
nucleolar protein 10 |
| chr23_-_35790235 | 0.38 |
ENSDART00000142369
ENSDART00000141141 ENSDART00000011004 |
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr11_+_17984167 | 0.37 |
ENSDART00000020283
ENSDART00000188329 |
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr18_+_13275735 | 0.37 |
ENSDART00000148127
|
plcg2
|
phospholipase C, gamma 2 |
| chr3_+_7808459 | 0.37 |
ENSDART00000162374
|
hook2
|
hook microtubule-tethering protein 2 |
| chr21_-_1640547 | 0.37 |
ENSDART00000151041
|
zgc:152948
|
zgc:152948 |
| chr11_+_17984354 | 0.37 |
ENSDART00000179986
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr22_+_508290 | 0.37 |
ENSDART00000135403
|
nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr8_-_39884359 | 0.36 |
ENSDART00000131372
|
mlec
|
malectin |
| chr21_-_36571804 | 0.36 |
ENSDART00000138129
|
wwc1
|
WW and C2 domain containing 1 |
| chr7_+_7696665 | 0.36 |
ENSDART00000091099
|
ino80b
|
INO80 complex subunit B |
| chr3_-_31158382 | 0.35 |
ENSDART00000076764
ENSDART00000076796 |
smg1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr10_+_11767791 | 0.34 |
ENSDART00000092047
|
ppwd1
|
peptidylprolyl isomerase domain and WD repeat containing 1 |
| chr17_-_25831569 | 0.34 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr8_+_36560019 | 0.34 |
ENSDART00000136418
ENSDART00000061378 ENSDART00000185237 |
sf3a1
|
splicing factor 3a, subunit 1 |
| chr4_-_5108844 | 0.33 |
ENSDART00000132666
ENSDART00000136096 |
tmem209
|
transmembrane protein 209 |
| chr6_+_12527725 | 0.33 |
ENSDART00000149328
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr4_+_57881965 | 0.31 |
ENSDART00000162234
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr16_+_40024883 | 0.31 |
ENSDART00000110100
|
hint3
|
histidine triad nucleotide binding protein 3 |
| chr4_+_76926059 | 0.31 |
ENSDART00000136192
|
si:dkey-240n22.6
|
si:dkey-240n22.6 |
| chr10_-_13343831 | 0.30 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr8_-_19246342 | 0.30 |
ENSDART00000147172
|
abhd17ab
|
abhydrolase domain containing 17Ab |
| chr13_+_35528607 | 0.30 |
ENSDART00000075414
ENSDART00000112947 |
wdr27
|
WD repeat domain 27 |
| chr23_+_4709607 | 0.30 |
ENSDART00000166503
ENSDART00000158752 ENSDART00000163860 ENSDART00000172739 |
raf1a
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a Raf-1 proto-oncogene, serine/threonine kinase a |
| chr19_-_5769553 | 0.29 |
ENSDART00000175003
|
si:ch211-264f5.6
|
si:ch211-264f5.6 |
| chr3_+_23092762 | 0.29 |
ENSDART00000142884
ENSDART00000024136 |
gngt2a
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2a |
| chr7_-_67248829 | 0.28 |
ENSDART00000192442
|
znf143a
|
zinc finger protein 143a |
| chr12_-_35944654 | 0.28 |
ENSDART00000162579
ENSDART00000164199 |
dnai2a
|
dynein, axonemal, intermediate chain 2a |
| chr4_+_2482046 | 0.28 |
ENSDART00000103371
|
zdhhc17
|
zinc finger, DHHC-type containing 17 |
| chr19_-_5769728 | 0.28 |
ENSDART00000133106
|
si:ch211-264f5.6
|
si:ch211-264f5.6 |
| chr12_-_4532066 | 0.28 |
ENSDART00000092687
|
trpm4b.2
|
transient receptor potential cation channel, subfamily M, member 4b, transient receptor potential cation channel, subfamily M, member 4b, tandem duplicate 2 |
| chr16_-_36748374 | 0.28 |
ENSDART00000133310
|
pik3r4
|
phosphoinositide-3-kinase, regulatory subunit 4 |
| chr18_+_35128685 | 0.27 |
ENSDART00000151579
|
si:ch211-195m9.3
|
si:ch211-195m9.3 |
| chr2_-_17392799 | 0.27 |
ENSDART00000136470
ENSDART00000141188 |
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr15_+_29140126 | 0.26 |
ENSDART00000060034
|
zgc:113149
|
zgc:113149 |
| chr10_-_11261565 | 0.26 |
ENSDART00000146727
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr1_-_51038885 | 0.26 |
ENSDART00000035150
|
spast
|
spastin |
| chr23_-_18913032 | 0.26 |
ENSDART00000136678
|
si:ch211-209j10.6
|
si:ch211-209j10.6 |
| chr2_-_17393216 | 0.26 |
ENSDART00000123137
|
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr4_-_11064073 | 0.26 |
ENSDART00000150760
|
si:dkey-21h14.8
|
si:dkey-21h14.8 |
| chr7_-_8712148 | 0.26 |
ENSDART00000065488
|
tex261
|
testis expressed 261 |
| chr9_-_32177117 | 0.24 |
ENSDART00000078568
|
sf3b1
|
splicing factor 3b, subunit 1 |
| chr22_-_24791505 | 0.24 |
ENSDART00000136837
|
vtg4
|
vitellogenin 4 |
| chr4_-_2219705 | 0.24 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr5_+_29851433 | 0.24 |
ENSDART00000143434
|
ubash3ba
|
ubiquitin associated and SH3 domain containing Ba |
| chr21_+_37090585 | 0.24 |
ENSDART00000182971
|
znf346
|
zinc finger protein 346 |
| chr3_+_38540411 | 0.23 |
ENSDART00000154943
|
si:dkey-7f16.3
|
si:dkey-7f16.3 |
| chr3_+_1637358 | 0.23 |
ENSDART00000180266
|
CR394546.5
|
|
| chr11_-_10456387 | 0.23 |
ENSDART00000011087
ENSDART00000081827 |
ect2
|
epithelial cell transforming 2 |
| chr15_-_26931541 | 0.23 |
ENSDART00000027563
|
ccdc9
|
coiled-coil domain containing 9 |
| chr16_+_4654333 | 0.23 |
ENSDART00000167665
|
LIN28A
|
si:ch1073-284b18.2 |
| chr1_+_27977297 | 0.23 |
ENSDART00000180692
ENSDART00000166819 |
sugt1
|
SGT1 homolog, MIS12 kinetochore complex assembly cochaperone |
| chr22_+_2751887 | 0.23 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr25_-_27621268 | 0.23 |
ENSDART00000146205
ENSDART00000073511 |
hyal6
|
hyaluronoglucosaminidase 6 |
| chr24_-_38110779 | 0.23 |
ENSDART00000147783
|
crp
|
c-reactive protein, pentraxin-related |
| chr5_+_32076109 | 0.23 |
ENSDART00000051357
ENSDART00000144510 |
zmat5
|
zinc finger, matrin-type 5 |
| chr11_+_45436703 | 0.23 |
ENSDART00000168295
ENSDART00000173293 |
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr15_-_2754056 | 0.23 |
ENSDART00000129380
|
ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr13_+_33268657 | 0.22 |
ENSDART00000002095
|
tmem39b
|
transmembrane protein 39B |
| chr15_+_857148 | 0.21 |
ENSDART00000156949
|
si:dkey-7i4.13
|
si:dkey-7i4.13 |
| chr16_+_13818743 | 0.21 |
ENSDART00000090191
|
flcn
|
folliculin |
| chr19_+_43885770 | 0.21 |
ENSDART00000135599
|
lypla2
|
lysophospholipase II |
| chr5_-_13251907 | 0.21 |
ENSDART00000176774
ENSDART00000030553 |
top3b
|
DNA topoisomerase III beta |
| chr3_+_40164129 | 0.20 |
ENSDART00000102526
|
gfer
|
growth factor, augmenter of liver regeneration (ERV1 homolog, S. cerevisiae) |
| chr24_+_9693951 | 0.20 |
ENSDART00000082411
|
topbp1
|
DNA topoisomerase II binding protein 1 |
| chr13_-_50366950 | 0.20 |
ENSDART00000098209
|
sirt1
|
sirtuin 1 |
| chr24_+_15020402 | 0.20 |
ENSDART00000148102
|
dok6
|
docking protein 6 |
| chr5_+_61459422 | 0.20 |
ENSDART00000050902
|
polr2j
|
polymerase (RNA) II (DNA directed) polypeptide J |
| chr12_+_47698356 | 0.20 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr24_+_28953089 | 0.20 |
ENSDART00000153761
|
rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr22_-_21676364 | 0.19 |
ENSDART00000183668
|
tle2b
|
transducin like enhancer of split 2b |
| chr8_+_29635968 | 0.18 |
ENSDART00000139029
ENSDART00000091409 |
smarcad1a
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 a |
| chr11_+_1602916 | 0.18 |
ENSDART00000184434
ENSDART00000112597 ENSDART00000192165 |
si:dkey-40c23.2
si:dkey-40c23.3
|
si:dkey-40c23.2 si:dkey-40c23.3 |
| chr18_+_20481982 | 0.18 |
ENSDART00000128139
|
kbtbd4
|
kelch repeat and BTB (POZ) domain containing 4 |
| chr1_-_42289704 | 0.17 |
ENSDART00000150124
|
si:ch211-71k14.1
|
si:ch211-71k14.1 |
| chr22_-_26865181 | 0.17 |
ENSDART00000138311
|
hmox2a
|
heme oxygenase 2a |
| chr5_+_19343880 | 0.17 |
ENSDART00000148130
|
acacb
|
acetyl-CoA carboxylase beta |
| chr7_+_26534131 | 0.17 |
ENSDART00000173980
|
si:dkey-62k3.5
|
si:dkey-62k3.5 |
| chr16_-_44945224 | 0.17 |
ENSDART00000156921
|
ncam3
|
neural cell adhesion molecule 3 |
| chr11_-_10456553 | 0.17 |
ENSDART00000169509
ENSDART00000185574 ENSDART00000188276 |
ect2
|
epithelial cell transforming 2 |
| chr8_+_31717175 | 0.17 |
ENSDART00000013434
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr4_-_43640507 | 0.17 |
ENSDART00000150700
|
si:dkey-29p23.2
|
si:dkey-29p23.2 |
| chr20_+_25586099 | 0.17 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr23_-_19140781 | 0.16 |
ENSDART00000143580
|
si:ch73-381f5.2
|
si:ch73-381f5.2 |
| chr4_+_33012407 | 0.16 |
ENSDART00000151873
|
si:dkey-26h11.2
|
si:dkey-26h11.2 |
| chr23_-_36418708 | 0.15 |
ENSDART00000132273
|
znf740b
|
zinc finger protein 740b |
| chr1_+_47499888 | 0.15 |
ENSDART00000027624
|
stn1
|
STN1, CST complex subunit |
| chr2_+_2169337 | 0.15 |
ENSDART00000179939
|
higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr1_+_49668423 | 0.15 |
ENSDART00000150880
|
tsga10
|
testis specific, 10 |
| chr6_-_12588044 | 0.15 |
ENSDART00000047896
|
slc15a1b
|
solute carrier family 15 (oligopeptide transporter), member 1b |
| chr1_+_32051581 | 0.15 |
ENSDART00000146602
|
sts
|
steroid sulfatase (microsomal), isozyme S |
| chr13_+_9468535 | 0.15 |
ENSDART00000135088
ENSDART00000164270 ENSDART00000099619 ENSDART00000164656 |
HTRA2 (1 of many)
|
si:dkey-265c15.6 |
| chr16_+_28728347 | 0.15 |
ENSDART00000149240
|
si:dkey-24i24.3
|
si:dkey-24i24.3 |
| chr13_+_646700 | 0.15 |
ENSDART00000006892
|
tp53bp2a
|
tumor protein p53 binding protein, 2a |
| chr8_+_31716872 | 0.15 |
ENSDART00000161121
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr21_-_26028205 | 0.14 |
ENSDART00000034875
|
sdf2
|
stromal cell-derived factor 2 |
| chr3_+_29179329 | 0.14 |
ENSDART00000085216
ENSDART00000190136 |
cacna1i
|
calcium channel, voltage-dependent, T type, alpha 1I subunit |
| chr25_-_25058508 | 0.14 |
ENSDART00000087570
ENSDART00000178891 |
FQ311928.1
|
|
| chr24_+_19415124 | 0.14 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr22_-_5252005 | 0.13 |
ENSDART00000132942
ENSDART00000081801 |
ncln
|
nicalin |
| chr4_-_46915962 | 0.13 |
ENSDART00000169555
|
si:ch211-134c10.1
|
si:ch211-134c10.1 |
| chr8_+_50953776 | 0.13 |
ENSDART00000013870
|
zgc:56596
|
zgc:56596 |
| chr17_+_20589553 | 0.13 |
ENSDART00000154447
|
si:ch73-288o11.4
|
si:ch73-288o11.4 |
| chr10_+_29850330 | 0.13 |
ENSDART00000168898
|
hspa8
|
heat shock protein 8 |
| chr13_+_15656042 | 0.13 |
ENSDART00000134240
|
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr4_-_63769637 | 0.13 |
ENSDART00000158757
|
znf1098
|
zinc finger protein 1098 |
| chr7_-_11596450 | 0.12 |
ENSDART00000173863
|
stard5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr4_-_43388943 | 0.12 |
ENSDART00000150796
|
si:dkey-29j8.2
|
si:dkey-29j8.2 |
| chr22_-_9183944 | 0.12 |
ENSDART00000188599
|
si:ch211-213a13.5
|
si:ch211-213a13.5 |
| chr16_-_27677930 | 0.12 |
ENSDART00000145991
|
tbrg4
|
transforming growth factor beta regulator 4 |
| chr23_-_16485190 | 0.12 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr5_+_38040407 | 0.12 |
ENSDART00000139936
|
si:dkey-111e8.4
|
si:dkey-111e8.4 |
| chr10_+_11261576 | 0.11 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr13_-_12602920 | 0.11 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr7_+_66884291 | 0.11 |
ENSDART00000187499
|
sbf2
|
SET binding factor 2 |
| chr10_+_22527715 | 0.11 |
ENSDART00000134864
|
gigyf1b
|
GRB10 interacting GYF protein 1b |
| chr7_+_66884570 | 0.11 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr7_-_50410524 | 0.11 |
ENSDART00000083346
|
hypk
|
huntingtin interacting protein K |
| chr20_+_11731039 | 0.10 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr6_+_7533601 | 0.10 |
ENSDART00000057823
|
pa2g4a
|
proliferation-associated 2G4, a |
| chr11_+_11267493 | 0.10 |
ENSDART00000148425
|
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr1_+_513986 | 0.10 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr8_-_45867358 | 0.10 |
ENSDART00000132810
|
adam9
|
ADAM metallopeptidase domain 9 |
| chr19_+_1688727 | 0.09 |
ENSDART00000115136
ENSDART00000166744 |
dennd3a
|
DENN/MADD domain containing 3a |
| chr17_-_29224908 | 0.09 |
ENSDART00000156288
|
si:dkey-28g23.6
|
si:dkey-28g23.6 |
| chr4_-_19921410 | 0.08 |
ENSDART00000184359
ENSDART00000179965 |
BX323820.1
|
|
| chr1_-_513762 | 0.08 |
ENSDART00000148162
ENSDART00000144606 |
trmt10c
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr2_+_19522082 | 0.08 |
ENSDART00000146098
|
pimr49
|
Pim proto-oncogene, serine/threonine kinase, related 49 |
| chr7_-_34448076 | 0.08 |
ENSDART00000170935
|
nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr24_+_21540842 | 0.08 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr6_+_40591149 | 0.08 |
ENSDART00000189060
ENSDART00000188298 |
frs3
|
fibroblast growth factor receptor substrate 3 |
| chr7_+_4474880 | 0.08 |
ENSDART00000143528
|
si:dkey-83f18.14
|
si:dkey-83f18.14 |
| chr17_+_26611929 | 0.08 |
ENSDART00000166450
ENSDART00000087023 |
ttc7b
|
tetratricopeptide repeat domain 7B |
| chr9_+_24065855 | 0.07 |
ENSDART00000161468
ENSDART00000171577 ENSDART00000172743 ENSDART00000159324 ENSDART00000079689 ENSDART00000023196 ENSDART00000101577 |
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr13_-_39736938 | 0.07 |
ENSDART00000141645
|
zgc:171482
|
zgc:171482 |
| chr12_-_28363111 | 0.07 |
ENSDART00000016283
ENSDART00000164156 |
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr20_-_34750363 | 0.07 |
ENSDART00000152845
|
znf395b
|
zinc finger protein 395b |
| chr13_-_9467944 | 0.07 |
ENSDART00000136582
|
pimr152
|
Pim proto-oncogene, serine/threonine kinase, related 152 |
| chr4_-_67980261 | 0.07 |
ENSDART00000182305
|
si:ch211-223k15.1
|
si:ch211-223k15.1 |
| chr8_-_36287046 | 0.07 |
ENSDART00000162877
|
si:busm1-194e12.11
|
si:busm1-194e12.11 |
| chr10_+_34047352 | 0.07 |
ENSDART00000169333
|
HTRA2 (1 of many)
|
si:dkey-10b15.8 |
| chr7_+_4657987 | 0.07 |
ENSDART00000144321
ENSDART00000142344 |
si:dkey-83f18.2
|
si:dkey-83f18.2 |
| chr16_-_41515662 | 0.07 |
ENSDART00000166201
ENSDART00000127243 |
siglec15l
|
sialic acid binding Ig-like lectin 15, like |
| chr6_-_52788213 | 0.06 |
ENSDART00000179880
|
rbpjl
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr1_-_22851481 | 0.06 |
ENSDART00000054386
|
qdprb1
|
quinoid dihydropteridine reductase b1 |
| chr25_+_29474982 | 0.06 |
ENSDART00000130410
|
il17rel
|
interleukin 17 receptor E-like |
| chr2_-_23603061 | 0.05 |
ENSDART00000132072
|
si:dkey-58b18.9
|
si:dkey-58b18.9 |
| chr16_-_21140097 | 0.05 |
ENSDART00000145837
ENSDART00000146500 |
si:dkey-271j15.3
|
si:dkey-271j15.3 |
| chr17_-_16422654 | 0.05 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr3_-_31254379 | 0.05 |
ENSDART00000189376
|
apnl
|
actinoporin-like protein |
| chr20_-_34750045 | 0.04 |
ENSDART00000186130
|
znf395b
|
zinc finger protein 395b |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc6a+hoxc6b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.2 | 0.5 | GO:0060945 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.1 | 0.6 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.1 | 0.7 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 0.5 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 0.5 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.3 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.1 | 0.5 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 0.5 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) |
| 0.1 | 0.3 | GO:0051230 | protein hexamerization(GO:0034214) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.1 | 0.5 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.1 | 0.3 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.4 | GO:0032475 | otolith formation(GO:0032475) |
| 0.1 | 0.2 | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902165) |
| 0.0 | 0.3 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 0.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.5 | GO:0016074 | snoRNA metabolic process(GO:0016074) |
| 0.0 | 0.2 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.5 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.2 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.4 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.9 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.2 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.4 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.1 | GO:1904478 | regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.2 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.0 | 0.7 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 1.2 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 1.6 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.4 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.2 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.1 | GO:1902946 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.0 | 0.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.2 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 0.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.1 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.1 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.2 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.1 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 0.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.1 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.1 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.4 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.1 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.0 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.2 | 0.5 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.4 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 0.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.3 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 0.4 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.2 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 0.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.7 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 1.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.0 | GO:0044217 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.1 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.4 | GO:0032040 | small-subunit processome(GO:0032040) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.2 | 0.5 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.1 | 0.4 | GO:0072591 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.1 | 0.2 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 1.0 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.5 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.1 | 0.9 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.3 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 0.5 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.6 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.2 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.5 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.2 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.5 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.7 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.5 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.3 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.2 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.1 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.0 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 1.7 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.4 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 0.6 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 0.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.6 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.4 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 1.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 0.7 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.2 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.4 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.4 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |