Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
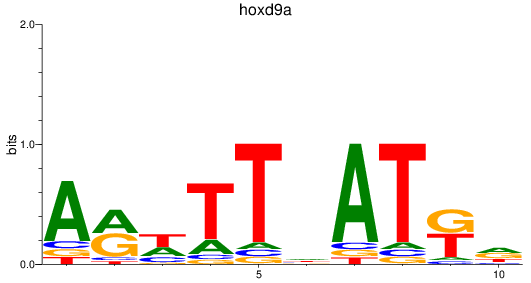
Results for hoxd9a
Z-value: 0.38
Transcription factors associated with hoxd9a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxd9a
|
ENSDARG00000059274 | homeobox D9a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxd9a | dr11_v1_chr9_-_1965727_1965727 | -0.02 | 9.6e-01 | Click! |
Activity profile of hoxd9a motif
Sorted Z-values of hoxd9a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_418932 | 0.16 |
ENSDART00000059193
|
prtgb
|
protogenin homolog b (Gallus gallus) |
| chr2_+_11685742 | 0.13 |
ENSDART00000138562
|
greb1l
|
growth regulation by estrogen in breast cancer-like |
| chr22_+_37888249 | 0.12 |
ENSDART00000076082
|
fetub
|
fetuin B |
| chr18_-_36066087 | 0.12 |
ENSDART00000059352
ENSDART00000145177 |
exosc5
|
exosome component 5 |
| chr3_-_55121125 | 0.10 |
ENSDART00000125092
|
hbae1
|
hemoglobin, alpha embryonic 1 |
| chr7_-_8374950 | 0.10 |
ENSDART00000057101
|
aep1
|
aerolysin-like protein |
| chr20_-_43741159 | 0.10 |
ENSDART00000192621
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr21_+_39963851 | 0.10 |
ENSDART00000144435
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr2_-_57837838 | 0.10 |
ENSDART00000010699
|
sf3a2
|
splicing factor 3a, subunit 2 |
| chr2_+_4402765 | 0.10 |
ENSDART00000159525
|
bambib
|
BMP and activin membrane-bound inhibitor homolog (Xenopus laevis) b |
| chr16_+_1383914 | 0.10 |
ENSDART00000185089
|
cers2b
|
ceramide synthase 2b |
| chr13_-_37620091 | 0.09 |
ENSDART00000135875
ENSDART00000193270 ENSDART00000018064 |
zgc:152791
|
zgc:152791 |
| chr5_+_43006422 | 0.09 |
ENSDART00000009182
|
aqp3a
|
aquaporin 3a |
| chr16_+_13883872 | 0.09 |
ENSDART00000101304
ENSDART00000136005 |
atg12
|
ATG12 autophagy related 12 homolog (S. cerevisiae) |
| chr22_-_24880824 | 0.09 |
ENSDART00000061165
|
vtg2
|
vitellogenin 2 |
| chr16_+_23913943 | 0.09 |
ENSDART00000175404
ENSDART00000129525 |
apoa4b.1
|
apolipoprotein A-IV b, tandem duplicate 1 |
| chr22_-_26945493 | 0.08 |
ENSDART00000077411
|
cxcl12b
|
chemokine (C-X-C motif) ligand 12b (stromal cell-derived factor 1) |
| chr20_-_8094718 | 0.08 |
ENSDART00000122902
|
si:ch211-232i5.3
|
si:ch211-232i5.3 |
| chr3_-_32817274 | 0.08 |
ENSDART00000142582
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr13_-_28674422 | 0.08 |
ENSDART00000122754
ENSDART00000057574 |
nt5c2a
|
5'-nucleotidase, cytosolic IIa |
| chr25_+_31276842 | 0.08 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr1_+_38362412 | 0.08 |
ENSDART00000075086
|
cep44
|
centrosomal protein 44 |
| chr3_+_15805917 | 0.08 |
ENSDART00000055834
|
phospho1
|
phosphatase, orphan 1 |
| chr13_+_48359573 | 0.08 |
ENSDART00000161959
ENSDART00000165311 |
msh6
|
mutS homolog 6 (E. coli) |
| chr19_+_10855158 | 0.07 |
ENSDART00000172219
ENSDART00000170826 |
apoea
|
apolipoprotein Ea |
| chr6_-_53426773 | 0.07 |
ENSDART00000162791
|
mst1
|
macrophage stimulating 1 |
| chr12_-_990149 | 0.07 |
ENSDART00000054367
|
kdelr2b
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2b |
| chr13_-_36535128 | 0.07 |
ENSDART00000043312
|
srsf5a
|
serine/arginine-rich splicing factor 5a |
| chr6_+_21684296 | 0.07 |
ENSDART00000057223
|
rhebl1
|
Ras homolog, mTORC1 binding like 1 |
| chr21_+_39941875 | 0.07 |
ENSDART00000190414
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr23_-_23401305 | 0.07 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr8_+_22359881 | 0.07 |
ENSDART00000187867
|
zgc:153631
|
zgc:153631 |
| chr12_+_1000323 | 0.07 |
ENSDART00000054363
|
si:ch1073-272o11.3
|
si:ch1073-272o11.3 |
| chr16_-_47427016 | 0.07 |
ENSDART00000074575
|
sept7b
|
septin 7b |
| chr14_-_35462148 | 0.07 |
ENSDART00000045809
|
srpx2
|
sushi-repeat containing protein, X-linked 2 |
| chr22_+_15959844 | 0.07 |
ENSDART00000182201
|
stil
|
scl/tal1 interrupting locus |
| chr17_-_14876758 | 0.06 |
ENSDART00000155857
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr15_-_14212777 | 0.06 |
ENSDART00000165572
|
CR925813.1
|
|
| chr2_-_33993533 | 0.06 |
ENSDART00000140910
ENSDART00000077304 |
ptch2
|
patched 2 |
| chr24_+_41690545 | 0.06 |
ENSDART00000160069
|
lama1
|
laminin, alpha 1 |
| chr21_+_39941184 | 0.06 |
ENSDART00000133604
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr5_-_46329880 | 0.06 |
ENSDART00000156577
|
si:ch211-130m23.5
|
si:ch211-130m23.5 |
| chr23_+_7379728 | 0.06 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr21_+_39941559 | 0.06 |
ENSDART00000189718
ENSDART00000160875 ENSDART00000135235 |
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr12_+_9499742 | 0.06 |
ENSDART00000044150
ENSDART00000136354 |
dnajc9
|
DnaJ (Hsp40) homolog, subfamily C, member 9 |
| chr16_+_54209504 | 0.06 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr9_+_48219111 | 0.06 |
ENSDART00000111225
ENSDART00000145972 |
ccdc173
|
coiled-coil domain containing 173 |
| chr18_+_33580391 | 0.06 |
ENSDART00000189300
|
si:dkey-47k20.4
|
si:dkey-47k20.4 |
| chr5_-_1963498 | 0.06 |
ENSDART00000073462
|
rplp0
|
ribosomal protein, large, P0 |
| chr9_-_303658 | 0.06 |
ENSDART00000160338
|
si:ch211-166e11.5
|
si:ch211-166e11.5 |
| chr14_-_7141550 | 0.06 |
ENSDART00000172020
|
trpt1
|
tRNA phosphotransferase 1 |
| chr24_-_38083378 | 0.06 |
ENSDART00000056381
|
crp2
|
C-reactive protein 2 |
| chr19_+_7835025 | 0.05 |
ENSDART00000026276
|
cks1b
|
CDC28 protein kinase regulatory subunit 1B |
| chr2_+_30739680 | 0.05 |
ENSDART00000101861
|
tcea1
|
transcription elongation factor A (SII), 1 |
| chr16_-_7443388 | 0.05 |
ENSDART00000017445
|
prdm1a
|
PR domain containing 1a, with ZNF domain |
| chr6_+_43400059 | 0.05 |
ENSDART00000143374
|
mitfa
|
microphthalmia-associated transcription factor a |
| chr22_+_5752257 | 0.05 |
ENSDART00000143052
|
cox14
|
COX14 cytochrome c oxidase assembly factor |
| chr3_-_31115996 | 0.05 |
ENSDART00000058955
|
arl6ip1
|
ADP-ribosylation factor-like 6 interacting protein 1 |
| chr1_-_338445 | 0.05 |
ENSDART00000010092
|
gas6
|
growth arrest-specific 6 |
| chr17_-_1174960 | 0.05 |
ENSDART00000161202
|
dnajc17
|
DnaJ (Hsp40) homolog, subfamily C, member 17 |
| chr22_-_22301929 | 0.05 |
ENSDART00000142027
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr20_+_26538137 | 0.05 |
ENSDART00000045397
|
stx11b.1
|
syntaxin 11b, tandem duplicate 1 |
| chr15_+_47440477 | 0.05 |
ENSDART00000002384
|
phox2a
|
paired-like homeobox 2a |
| chr14_+_46342882 | 0.05 |
ENSDART00000193707
ENSDART00000060577 |
tmem33
|
transmembrane protein 33 |
| chr12_+_13164706 | 0.05 |
ENSDART00000112565
|
paqr4b
|
progestin and adipoQ receptor family member IVb |
| chr21_+_21357309 | 0.05 |
ENSDART00000079634
ENSDART00000146102 |
kcnk12l
|
potassium channel, subfamily K, member 12 like |
| chr20_+_36806398 | 0.05 |
ENSDART00000153317
|
abracl
|
ABRA C-terminal like |
| chr8_+_20393250 | 0.05 |
ENSDART00000015038
|
zap70
|
zeta chain of T cell receptor associated protein kinase 70 |
| chr3_-_39695856 | 0.04 |
ENSDART00000148247
|
b9d1
|
B9 protein domain 1 |
| chr5_-_31897139 | 0.04 |
ENSDART00000191957
|
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr22_+_15960005 | 0.04 |
ENSDART00000033617
|
stil
|
scl/tal1 interrupting locus |
| chr20_-_35076387 | 0.04 |
ENSDART00000019196
|
lft1
|
lefty1 |
| chr2_+_10878406 | 0.04 |
ENSDART00000091497
|
tceanc2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chr12_+_19199735 | 0.04 |
ENSDART00000066393
|
pdap1a
|
pdgfa associated protein 1a |
| chr6_-_8377055 | 0.04 |
ENSDART00000131513
|
ilf3a
|
interleukin enhancer binding factor 3a |
| chr5_-_12093618 | 0.04 |
ENSDART00000161542
|
lrrc74b
|
leucine rich repeat containing 74B |
| chr5_+_23083555 | 0.04 |
ENSDART00000159562
|
zdhhc15a
|
zinc finger, DHHC-type containing 15a |
| chr23_+_9035578 | 0.04 |
ENSDART00000140455
|
ccm2l
|
cerebral cavernous malformation 2-like |
| chr12_+_24952902 | 0.04 |
ENSDART00000189086
ENSDART00000014868 |
calm3a
|
calmodulin 3a (phosphorylase kinase, delta) |
| chr13_+_39236141 | 0.04 |
ENSDART00000111458
|
zgc:172136
|
zgc:172136 |
| chr6_+_41191482 | 0.04 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr21_+_3166519 | 0.04 |
ENSDART00000160585
|
hnrnpabb
|
heterogeneous nuclear ribonucleoprotein A/Bb |
| chr4_+_71401321 | 0.04 |
ENSDART00000158076
|
si:ch211-76m11.7
|
si:ch211-76m11.7 |
| chr15_-_34322915 | 0.04 |
ENSDART00000190543
ENSDART00000019651 ENSDART00000193601 |
dgkb
|
diacylglycerol kinase, beta |
| chr18_+_9810642 | 0.04 |
ENSDART00000143965
|
CACNA2D1
|
si:dkey-266j7.2 |
| chr15_-_1844048 | 0.04 |
ENSDART00000102410
|
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr19_-_37508571 | 0.04 |
ENSDART00000018255
|
ilf2
|
interleukin enhancer binding factor 2 |
| chr22_-_10165446 | 0.04 |
ENSDART00000142012
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr10_+_23537576 | 0.04 |
ENSDART00000130515
|
CR847844.1
|
|
| chr3_+_22442445 | 0.04 |
ENSDART00000190921
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr4_+_14899720 | 0.04 |
ENSDART00000187456
|
akr1b1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr3_+_13848226 | 0.04 |
ENSDART00000184342
|
ilf3b
|
interleukin enhancer binding factor 3b |
| chr1_-_22851481 | 0.04 |
ENSDART00000054386
|
qdprb1
|
quinoid dihydropteridine reductase b1 |
| chr7_-_26306546 | 0.04 |
ENSDART00000140817
|
zgc:77439
|
zgc:77439 |
| chr13_-_41155472 | 0.04 |
ENSDART00000160588
|
si:dkeyp-86d6.2
|
si:dkeyp-86d6.2 |
| chr16_-_47426482 | 0.04 |
ENSDART00000148631
ENSDART00000149723 |
sept7b
|
septin 7b |
| chr3_-_9444749 | 0.04 |
ENSDART00000182191
|
FO904885.3
|
|
| chr15_+_846768 | 0.04 |
ENSDART00000155633
ENSDART00000191235 |
si:dkey-7i4.5
si:dkey-77f5.8
|
si:dkey-7i4.5 si:dkey-77f5.8 |
| chr3_+_46425262 | 0.04 |
ENSDART00000153690
|
sc:d0202
|
sc:d0202 |
| chr4_+_14900042 | 0.03 |
ENSDART00000018261
|
akr1b1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr2_-_10877765 | 0.03 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr10_-_24868581 | 0.03 |
ENSDART00000193055
|
CABZ01058107.1
|
|
| chr11_+_24815927 | 0.03 |
ENSDART00000146838
|
rabif
|
RAB interacting factor |
| chr22_+_20710434 | 0.03 |
ENSDART00000135521
|
peak3
|
PEAK family member 3 |
| chr14_+_45406299 | 0.03 |
ENSDART00000173142
ENSDART00000112377 |
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr21_+_43172506 | 0.03 |
ENSDART00000121725
|
zcchc10
|
zinc finger, CCHC domain containing 10 |
| chr19_+_6914095 | 0.03 |
ENSDART00000104906
|
si:ch73-367f21.4
|
si:ch73-367f21.4 |
| chr18_-_3542435 | 0.03 |
ENSDART00000184017
|
dcun1d5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) |
| chr11_+_45388126 | 0.03 |
ENSDART00000167826
ENSDART00000171274 |
srsf11
|
serine/arginine-rich splicing factor 11 |
| chr16_-_42461263 | 0.03 |
ENSDART00000109259
|
smarcc1a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1a |
| chr25_-_4192637 | 0.03 |
ENSDART00000153832
|
ppp1r32
|
protein phosphatase 1, regulatory subunit 32 |
| chr24_-_3477103 | 0.03 |
ENSDART00000143723
|
idi1
|
isopentenyl-diphosphate delta isomerase 1 |
| chr6_-_34938678 | 0.03 |
ENSDART00000186689
ENSDART00000131610 |
serbp1a
|
SERPINE1 mRNA binding protein 1a |
| chr2_-_48196092 | 0.03 |
ENSDART00000139944
|
snorc
|
secondary ossification center associated regulator of chondrocyte maturation |
| chr8_+_21133307 | 0.03 |
ENSDART00000056405
|
magoh
|
mago homolog, exon junction complex core component |
| chr2_+_19195841 | 0.03 |
ENSDART00000163137
ENSDART00000161095 |
elovl1a
|
ELOVL fatty acid elongase 1a |
| chr3_-_31115601 | 0.03 |
ENSDART00000139090
|
arl6ip1
|
ADP-ribosylation factor-like 6 interacting protein 1 |
| chr22_-_392224 | 0.03 |
ENSDART00000124720
|
CABZ01072989.1
|
|
| chr7_-_24046999 | 0.03 |
ENSDART00000144616
ENSDART00000124653 ENSDART00000127813 |
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr11_+_37201483 | 0.03 |
ENSDART00000160930
ENSDART00000173439 ENSDART00000171273 |
zgc:112265
|
zgc:112265 |
| chr22_+_2403068 | 0.03 |
ENSDART00000132925
ENSDART00000132569 |
zgc:112977
|
zgc:112977 |
| chr3_-_49382896 | 0.03 |
ENSDART00000169115
|
si:ch73-167f10.1
|
si:ch73-167f10.1 |
| chr9_-_12269847 | 0.03 |
ENSDART00000136558
ENSDART00000144734 ENSDART00000131766 ENSDART00000032344 |
nup35
|
nucleoporin 35 |
| chr9_+_21259820 | 0.03 |
ENSDART00000137024
ENSDART00000132324 |
ska3
|
spindle and kinetochore associated complex subunit 3 |
| chr10_-_40557210 | 0.03 |
ENSDART00000135297
|
taar18b
|
trace amine associated receptor 18b |
| chr5_-_61787969 | 0.03 |
ENSDART00000112744
|
gas2l2
|
growth arrest specific 2 like 2 |
| chr9_+_2333550 | 0.03 |
ENSDART00000016417
|
atp5mc3a
|
ATP synthase membrane subunit c locus 3a |
| chr14_+_26224731 | 0.03 |
ENSDART00000168673
|
gm2a
|
GM2 ganglioside activator |
| chr6_-_58764672 | 0.03 |
ENSDART00000154322
|
soat2
|
sterol O-acyltransferase 2 |
| chr15_+_37589698 | 0.03 |
ENSDART00000076066
ENSDART00000153894 ENSDART00000156298 |
lin37
|
lin-37 DREAM MuvB core complex component |
| chr23_+_4483083 | 0.03 |
ENSDART00000092389
|
nup210
|
nucleoporin 210 |
| chr1_-_69444 | 0.03 |
ENSDART00000166954
|
si:zfos-1011f11.1
|
si:zfos-1011f11.1 |
| chr21_+_45717930 | 0.03 |
ENSDART00000164315
|
ddx46
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 |
| chr23_+_36460239 | 0.03 |
ENSDART00000172441
|
lima1a
|
LIM domain and actin binding 1a |
| chr23_-_36446307 | 0.03 |
ENSDART00000136623
|
zgc:174906
|
zgc:174906 |
| chr8_-_36327328 | 0.03 |
ENSDART00000183333
|
zgc:103700
|
zgc:103700 |
| chr12_+_30723778 | 0.03 |
ENSDART00000153291
|
slc35f3a
|
solute carrier family 35, member F3a |
| chr20_+_1996202 | 0.03 |
ENSDART00000184143
|
CABZ01092781.1
|
|
| chr14_+_30795559 | 0.03 |
ENSDART00000006132
|
cfl1
|
cofilin 1 |
| chr2_+_25124498 | 0.03 |
ENSDART00000056801
|
msl2a
|
male-specific lethal 2 homolog a (Drosophila) |
| chr4_-_73739119 | 0.03 |
ENSDART00000108669
|
zgc:171551
|
zgc:171551 |
| chr4_-_1776352 | 0.03 |
ENSDART00000123089
|
dnajc2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr19_+_7810028 | 0.03 |
ENSDART00000081592
ENSDART00000140719 |
aqp10b
|
aquaporin 10b |
| chr19_-_42291355 | 0.03 |
ENSDART00000150919
ENSDART00000151034 |
si:ch211-191i18.2
|
si:ch211-191i18.2 |
| chr6_-_7776612 | 0.03 |
ENSDART00000190269
|
myh9a
|
myosin, heavy chain 9a, non-muscle |
| chr4_+_28473223 | 0.03 |
ENSDART00000150438
|
si:ch73-289h5.4
|
si:ch73-289h5.4 |
| chr17_-_13007484 | 0.03 |
ENSDART00000156812
|
si:dkeyp-33b5.4
|
si:dkeyp-33b5.4 |
| chr21_-_25213616 | 0.03 |
ENSDART00000122513
|
rfc2
|
replication factor C (activator 1) 2 |
| chr8_-_13064330 | 0.03 |
ENSDART00000129168
|
si:dkey-208b23.5
|
si:dkey-208b23.5 |
| chr22_+_6084771 | 0.03 |
ENSDART00000009590
|
si:dkey-19a16.16
|
si:dkey-19a16.16 |
| chr22_-_11661724 | 0.03 |
ENSDART00000025198
|
mettl21a
|
methyltransferase like 21A |
| chr3_+_39579393 | 0.03 |
ENSDART00000055170
|
cln3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr20_+_11731039 | 0.03 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr8_+_471342 | 0.03 |
ENSDART00000167205
|
nudt12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
| chr8_-_13029297 | 0.03 |
ENSDART00000144305
|
dennd2da
|
DENN/MADD domain containing 2Da |
| chr16_+_26706519 | 0.03 |
ENSDART00000142706
|
virma
|
vir like m6A methyltransferase associated |
| chr5_-_23179319 | 0.03 |
ENSDART00000161883
ENSDART00000136260 |
si:dkey-114c15.5
|
si:dkey-114c15.5 |
| chr15_-_32383340 | 0.03 |
ENSDART00000185632
|
c4
|
complement component 4 |
| chr22_-_11714104 | 0.03 |
ENSDART00000145265
ENSDART00000063127 ENSDART00000183743 |
crygs1
|
crystallin, gamma S1 |
| chr21_-_11646878 | 0.03 |
ENSDART00000162426
ENSDART00000135937 ENSDART00000131448 ENSDART00000148097 ENSDART00000133443 |
cast
|
calpastatin |
| chr10_+_11355841 | 0.03 |
ENSDART00000193067
ENSDART00000064215 |
cops4
|
COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis) |
| chr23_-_7052362 | 0.03 |
ENSDART00000127702
ENSDART00000192468 |
prpf6
|
PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae) |
| chr8_-_36287046 | 0.03 |
ENSDART00000162877
|
si:busm1-194e12.11
|
si:busm1-194e12.11 |
| chr6_-_52675630 | 0.03 |
ENSDART00000083830
|
sdc4
|
syndecan 4 |
| chr6_-_8392104 | 0.03 |
ENSDART00000081561
ENSDART00000181178 |
ilf3a
|
interleukin enhancer binding factor 3a |
| chr14_-_7137808 | 0.02 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr5_+_28830643 | 0.02 |
ENSDART00000051448
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr8_+_15251448 | 0.02 |
ENSDART00000063717
|
zgc:171480
|
zgc:171480 |
| chr15_-_3956906 | 0.02 |
ENSDART00000171202
|
clrn1
|
clarin 1 |
| chr22_+_31059919 | 0.02 |
ENSDART00000077063
|
sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr13_+_2523032 | 0.02 |
ENSDART00000172261
|
lhb
|
luteinizing hormone, beta polypeptide |
| chr10_-_36808348 | 0.02 |
ENSDART00000099320
|
dhrs13a.1
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 1 |
| chr11_-_42106310 | 0.02 |
ENSDART00000164635
|
si:ch211-193l2.10
|
si:ch211-193l2.10 |
| chr1_-_10647307 | 0.02 |
ENSDART00000103548
|
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr5_+_57480014 | 0.02 |
ENSDART00000135520
|
si:ch211-202f5.3
|
si:ch211-202f5.3 |
| chr9_+_52398531 | 0.02 |
ENSDART00000126215
|
dap1b
|
death associated protein 1b |
| chr14_+_32430982 | 0.02 |
ENSDART00000017179
ENSDART00000123382 ENSDART00000075593 |
f9a
|
coagulation factor IXa |
| chr1_+_41898919 | 0.02 |
ENSDART00000140845
|
si:dkey-37g12.1
|
si:dkey-37g12.1 |
| chr7_-_1148539 | 0.02 |
ENSDART00000173447
|
si:dkey-31g16.2
|
si:dkey-31g16.2 |
| chr12_+_2428247 | 0.02 |
ENSDART00000152529
|
lrrc18b
|
leucine rich repeat containing 18b |
| chr13_+_2894536 | 0.02 |
ENSDART00000183678
|
CABZ01087629.1
|
|
| chr5_-_23805387 | 0.02 |
ENSDART00000133805
|
gbgt1l4
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 4 |
| chr20_+_34792166 | 0.02 |
ENSDART00000061523
|
il17a/f3
|
interleukin 17a/f3 |
| chr8_+_7315625 | 0.02 |
ENSDART00000135655
ENSDART00000181048 |
selenoh
|
selenoprotein H |
| chr20_+_42978499 | 0.02 |
ENSDART00000138793
|
si:ch211-203k16.3
|
si:ch211-203k16.3 |
| chr19_+_10558535 | 0.02 |
ENSDART00000141292
|
usf2
|
upstream transcription factor 2, c-fos interacting |
| chr10_+_38708099 | 0.02 |
ENSDART00000172306
|
tmprss2
|
transmembrane protease, serine 2 |
| chr18_-_20532339 | 0.02 |
ENSDART00000060291
|
ighmbp2
|
immunoglobulin mu binding protein 2 |
| chr9_+_54679221 | 0.02 |
ENSDART00000167769
|
egfl6
|
EGF-like-domain, multiple 6 |
| chr25_+_20715950 | 0.02 |
ENSDART00000180223
|
ergic2
|
ERGIC and golgi 2 |
| chr20_+_53368611 | 0.02 |
ENSDART00000060432
|
cdc40
|
cell division cycle 40 homolog (S. cerevisiae) |
| chr16_-_16761164 | 0.02 |
ENSDART00000135872
|
si:dkey-27n14.1
|
si:dkey-27n14.1 |
| chr11_-_3366782 | 0.02 |
ENSDART00000127982
|
suox
|
sulfite oxidase |
| chr25_+_25438322 | 0.02 |
ENSDART00000150364
|
LRRC56
|
si:ch211-103e16.5 |
| chr16_-_31505802 | 0.02 |
ENSDART00000147616
|
si:ch211-251p5.5
|
si:ch211-251p5.5 |
| chr22_+_18188045 | 0.02 |
ENSDART00000140106
|
mef2b
|
myocyte enhancer factor 2b |
| chr20_+_1196386 | 0.02 |
ENSDART00000041192
|
lyrm2
|
LYR motif containing 2 |
| chr18_-_27897217 | 0.02 |
ENSDART00000175259
|
iqcg
|
IQ motif containing G |
| chr2_-_4868002 | 0.02 |
ENSDART00000189835
|
tfr1a
|
transferrin receptor 1a |
| chr10_+_24684799 | 0.02 |
ENSDART00000113696
|
thsd1
|
thrombospondin, type I, domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxd9a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.1 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:1900221 | regulation of nitric-oxide synthase activity(GO:0050999) positive regulation of nitric-oxide synthase activity(GO:0051000) beta-amyloid clearance(GO:0097242) regulation of beta-amyloid clearance(GO:1900221) |
| 0.0 | 0.1 | GO:0050764 | regulation of phagocytosis(GO:0050764) |
| 0.0 | 0.1 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.0 | 0.1 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.0 | 0.1 | GO:0000177 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.1 | GO:0032357 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0015105 | arsenite transmembrane transporter activity(GO:0015105) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.3 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |