Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
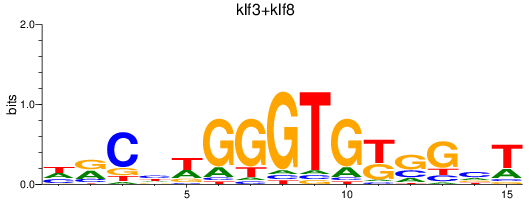
Results for klf3+klf8
Z-value: 1.18
Transcription factors associated with klf3+klf8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
klf3
|
ENSDARG00000015495 | Kruppel-like factor 3 (basic) |
|
klf8
|
ENSDARG00000060661 | Kruppel-like factor 8 |
|
klf3
|
ENSDARG00000114143 | Kruppel-like factor 3 (basic) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| klf8 | dr11_v1_chr21_+_38073210_38073216 | -0.93 | 2.9e-04 | Click! |
| klf3 | dr11_v1_chr1_-_18615063_18615063 | 0.84 | 4.4e-03 | Click! |
Activity profile of klf3+klf8 motif
Sorted Z-values of klf3+klf8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_2039060 | 3.36 |
ENSDART00000173879
|
si:cabz01007794.1
|
si:cabz01007794.1 |
| chr2_-_35566938 | 3.35 |
ENSDART00000029006
ENSDART00000077178 ENSDART00000125298 |
tnn
|
tenascin N |
| chr10_-_22797959 | 3.29 |
ENSDART00000183269
|
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr15_+_29085955 | 3.01 |
ENSDART00000156799
|
si:ch211-137a8.4
|
si:ch211-137a8.4 |
| chr16_+_23978978 | 2.85 |
ENSDART00000058964
ENSDART00000135084 |
apoa2
|
apolipoprotein A-II |
| chr1_-_44940830 | 2.84 |
ENSDART00000097500
ENSDART00000134464 ENSDART00000137216 |
tmem176
|
transmembrane protein 176 |
| chr18_-_46010 | 2.48 |
ENSDART00000052641
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr13_+_681628 | 2.25 |
ENSDART00000016604
|
abcc2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr14_+_13453130 | 2.07 |
ENSDART00000054849
|
pls3
|
plastin 3 (T isoform) |
| chr24_-_40667800 | 2.06 |
ENSDART00000169315
|
smyhc1
|
slow myosin heavy chain 1 |
| chr4_-_193762 | 2.04 |
ENSDART00000169667
|
ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr24_-_40668208 | 1.88 |
ENSDART00000171543
|
smyhc1
|
slow myosin heavy chain 1 |
| chr1_-_625875 | 1.70 |
ENSDART00000167331
|
appa
|
amyloid beta (A4) precursor protein a |
| chr13_-_992458 | 1.52 |
ENSDART00000114655
|
BFSP1
|
beaded filament structural protein 1 |
| chr4_-_191736 | 1.39 |
ENSDART00000169187
ENSDART00000192054 |
ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr1_+_44941031 | 1.31 |
ENSDART00000141145
|
si:dkey-9i23.16
|
si:dkey-9i23.16 |
| chr6_-_57655030 | 1.23 |
ENSDART00000155438
|
cbfa2t2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr19_+_56351 | 1.15 |
ENSDART00000168334
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr10_+_4875262 | 1.12 |
ENSDART00000165942
|
palm2
|
paralemmin 2 |
| chr7_+_35229645 | 1.12 |
ENSDART00000144327
|
tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr18_+_54354 | 1.05 |
ENSDART00000097163
|
zgc:158482
|
zgc:158482 |
| chr3_-_32927516 | 1.04 |
ENSDART00000140117
|
aoc2
|
amine oxidase, copper containing 2 |
| chr6_+_120181 | 0.99 |
ENSDART00000151209
ENSDART00000185930 |
cdkn2d
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr19_+_19786117 | 0.94 |
ENSDART00000167757
ENSDART00000163546 |
hoxa1a
|
homeobox A1a |
| chr6_+_41181869 | 0.93 |
ENSDART00000002046
|
opn1mw1
|
opsin 1 (cone pigments), medium-wave-sensitive, 1 |
| chr1_-_49434798 | 0.92 |
ENSDART00000150842
ENSDART00000150789 |
ATP5MD
|
si:dkeyp-80c12.10 |
| chr3_-_1190132 | 0.91 |
ENSDART00000149709
|
smdt1a
|
single-pass membrane protein with aspartate-rich tail 1a |
| chr10_+_4874542 | 0.91 |
ENSDART00000101431
|
palm2
|
paralemmin 2 |
| chr21_-_35832548 | 0.89 |
ENSDART00000180840
|
sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr19_+_233143 | 0.88 |
ENSDART00000175273
|
syngap1a
|
synaptic Ras GTPase activating protein 1a |
| chr2_-_30135446 | 0.86 |
ENSDART00000141906
|
trpa1a
|
transient receptor potential cation channel, subfamily A, member 1a |
| chr19_-_677713 | 0.85 |
ENSDART00000025146
|
slc6a19a.1
|
solute carrier family 6 (neutral amino acid transporter), member 19a, tandem duplicate 1 |
| chr5_-_64213253 | 0.85 |
ENSDART00000171711
|
gpsm1a
|
G protein signaling modulator 1a |
| chr21_+_44857293 | 0.82 |
ENSDART00000134365
ENSDART00000168217 ENSDART00000065083 |
fstl4
|
follistatin-like 4 |
| chr24_+_81527 | 0.82 |
ENSDART00000192139
|
reck
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr24_-_14212521 | 0.79 |
ENSDART00000130825
|
xkr9
|
XK, Kell blood group complex subunit-related family, member 9 |
| chr25_+_23489979 | 0.77 |
ENSDART00000158200
|
CU024870.1
|
|
| chr19_+_164013 | 0.74 |
ENSDART00000167379
|
carmil1
|
capping protein regulator and myosin 1 linker 1 |
| chr16_+_44768361 | 0.72 |
ENSDART00000036302
|
upk1a
|
uroplakin 1a |
| chr21_+_17542473 | 0.72 |
ENSDART00000005750
ENSDART00000141326 |
stom
|
stomatin |
| chr14_-_21218891 | 0.67 |
ENSDART00000158294
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr11_+_37278457 | 0.66 |
ENSDART00000188946
|
creld1a
|
cysteine-rich with EGF-like domains 1a |
| chr7_+_35229805 | 0.62 |
ENSDART00000173911
|
tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr23_-_45049044 | 0.62 |
ENSDART00000067630
|
SH3GL2 (1 of many)
|
SH3 domain containing GRB2 like 2, endophilin A1 |
| chr3_-_15444396 | 0.60 |
ENSDART00000104361
|
si:dkey-56d12.4
|
si:dkey-56d12.4 |
| chr5_-_69041102 | 0.59 |
ENSDART00000161561
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr20_-_54377933 | 0.59 |
ENSDART00000182664
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr9_-_11551608 | 0.58 |
ENSDART00000128629
|
fev
|
FEV (ETS oncogene family) |
| chr1_-_59571758 | 0.58 |
ENSDART00000193546
ENSDART00000167087 |
wfikkn1
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 1 |
| chr8_+_28065803 | 0.57 |
ENSDART00000178481
|
kcnd3
|
potassium voltage-gated channel, Shal-related subfamily, member 3 |
| chr6_+_4084739 | 0.56 |
ENSDART00000087661
|
sema5bb
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Bb |
| chr18_-_45617146 | 0.55 |
ENSDART00000146543
|
wt1b
|
wilms tumor 1b |
| chr9_-_12760323 | 0.54 |
ENSDART00000146265
|
myo10l3
|
myosin X-like 3 |
| chr20_-_52902693 | 0.52 |
ENSDART00000166115
ENSDART00000161050 |
ctsbb
|
cathepsin Bb |
| chr13_-_40238813 | 0.52 |
ENSDART00000044963
|
loxl4
|
lysyl oxidase-like 4 |
| chr18_-_40508528 | 0.52 |
ENSDART00000185249
|
chrna5
|
cholinergic receptor, nicotinic, alpha 5 |
| chr7_-_19600181 | 0.48 |
ENSDART00000100757
|
oxa1l
|
oxidase (cytochrome c) assembly 1-like |
| chr5_-_29587351 | 0.47 |
ENSDART00000136446
ENSDART00000051434 |
entpd2a.1
|
ectonucleoside triphosphate diphosphohydrolase 2a, tandem duplicate 1 |
| chr8_+_54013199 | 0.46 |
ENSDART00000158497
|
CABZ01079663.1
|
|
| chr21_+_27448856 | 0.45 |
ENSDART00000100784
|
cfbl
|
complement factor b-like |
| chr23_-_45439903 | 0.45 |
ENSDART00000170729
|
NPNT (1 of many)
|
nephronectin |
| chr21_+_11244068 | 0.45 |
ENSDART00000163432
|
arid6
|
AT-rich interaction domain 6 |
| chr5_-_24174956 | 0.45 |
ENSDART00000137631
|
si:ch211-137i24.10
|
si:ch211-137i24.10 |
| chr23_-_1017605 | 0.45 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr16_-_20492799 | 0.42 |
ENSDART00000014769
|
chn2
|
chimerin 2 |
| chr8_-_34762163 | 0.41 |
ENSDART00000114080
|
setd1bb
|
SET domain containing 1B, b |
| chr10_-_20637098 | 0.38 |
ENSDART00000080391
|
sprn2
|
shadow of prion protein 2 |
| chr25_+_37348730 | 0.37 |
ENSDART00000156639
|
pamr1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr23_+_9057999 | 0.37 |
ENSDART00000091899
|
ccm2l
|
cerebral cavernous malformation 2-like |
| chr3_+_55093581 | 0.36 |
ENSDART00000101709
|
hbaa2
|
hemoglobin, alpha adult 2 |
| chr1_+_57235896 | 0.35 |
ENSDART00000152621
|
si:dkey-27j5.7
|
si:dkey-27j5.7 |
| chr18_-_24988645 | 0.35 |
ENSDART00000136434
ENSDART00000085735 |
chd2
|
chromodomain helicase DNA binding protein 2 |
| chr23_-_1017428 | 0.34 |
ENSDART00000110588
ENSDART00000183158 |
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr20_+_54356272 | 0.31 |
ENSDART00000145735
|
znf410
|
zinc finger protein 410 |
| chr7_-_67316903 | 0.31 |
ENSDART00000159408
|
psmd7
|
proteasome 26S subunit, non-ATPase 7 |
| chr4_+_54618332 | 0.30 |
ENSDART00000171824
|
si:ch211-227e10.2
|
si:ch211-227e10.2 |
| chr6_-_52757276 | 0.30 |
ENSDART00000159154
|
AL954861.2
|
|
| chr4_+_50401133 | 0.29 |
ENSDART00000150644
|
si:dkey-156k2.7
|
si:dkey-156k2.7 |
| chr6_-_39344259 | 0.28 |
ENSDART00000104074
|
zgc:158846
|
zgc:158846 |
| chr23_-_44233408 | 0.27 |
ENSDART00000149318
ENSDART00000085528 |
zgc:158659
|
zgc:158659 |
| chr12_+_34763795 | 0.26 |
ENSDART00000153097
|
slc38a10
|
solute carrier family 38, member 10 |
| chr21_-_280769 | 0.26 |
ENSDART00000157753
|
plgrkt
|
plasminogen receptor, C-terminal lysine transmembrane protein |
| chr12_+_19030391 | 0.25 |
ENSDART00000153927
|
si:ch73-139e5.2
|
si:ch73-139e5.2 |
| chr25_+_16731802 | 0.25 |
ENSDART00000188715
|
BX004976.1
|
|
| chr8_-_44868020 | 0.24 |
ENSDART00000142712
|
cacna1fa
|
calcium channel, voltage-dependent, L type, alpha 1F subunit a |
| chr10_+_15106808 | 0.22 |
ENSDART00000141588
|
scpp8
|
secretory calcium-binding phosphoprotein 8 |
| chr6_+_41186320 | 0.22 |
ENSDART00000025241
|
opn1mw2
|
opsin 1 (cone pigments), medium-wave-sensitive, 2 |
| chr4_+_53976731 | 0.22 |
ENSDART00000165813
|
si:ch211-249c2.1
|
si:ch211-249c2.1 |
| chr7_-_32599669 | 0.22 |
ENSDART00000173752
|
kcna4
|
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr1_+_36194761 | 0.21 |
ENSDART00000053773
ENSDART00000147458 |
lsm6
|
LSM6 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr19_-_24218942 | 0.21 |
ENSDART00000189198
|
BX547993.2
|
|
| chr12_+_48851381 | 0.21 |
ENSDART00000187241
ENSDART00000187796 |
dlg5b.1
|
discs, large homolog 5b (Drosophila), tandem duplicate 1 |
| chr8_-_1973220 | 0.18 |
ENSDART00000184435
|
si:dkey-178e17.1
|
si:dkey-178e17.1 |
| chr7_-_67317119 | 0.18 |
ENSDART00000183915
|
psmd7
|
proteasome 26S subunit, non-ATPase 7 |
| chr8_-_30204650 | 0.16 |
ENSDART00000133209
|
zgc:162939
|
zgc:162939 |
| chr21_-_29197700 | 0.13 |
ENSDART00000172614
|
si:ch211-57b15.2
|
si:ch211-57b15.2 |
| chr18_+_5549672 | 0.12 |
ENSDART00000184970
|
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr7_-_6351021 | 0.12 |
ENSDART00000159542
|
zgc:112234
|
zgc:112234 |
| chr14_-_48260744 | 0.12 |
ENSDART00000183955
|
rxfp1
|
relaxin/insulin-like family peptide receptor 1 |
| chr1_-_53407087 | 0.12 |
ENSDART00000157910
|
elmod2
|
ELMO/CED-12 domain containing 2 |
| chr22_-_10019768 | 0.11 |
ENSDART00000162077
|
BX324216.2
|
|
| chr18_+_814328 | 0.10 |
ENSDART00000157801
ENSDART00000188492 ENSDART00000184489 |
fam219b
|
family with sequence similarity 219, member B |
| chr5_-_35953472 | 0.08 |
ENSDART00000143448
|
rxfp2l
|
relaxin/insulin-like family peptide receptor 2, like |
| chr19_+_7847920 | 0.08 |
ENSDART00000132454
|
si:dkeyp-85e10.1
|
si:dkeyp-85e10.1 |
| chr22_-_30948457 | 0.07 |
ENSDART00000139887
|
ssuh2.1
|
ssu-2 homolog, tandem duplicate 1 |
| chr2_-_16217344 | 0.06 |
ENSDART00000152031
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr15_-_47822597 | 0.05 |
ENSDART00000193236
ENSDART00000161391 |
CZQB01095947.1
|
|
| chr18_+_7309150 | 0.04 |
ENSDART00000052792
|
cers3b
|
ceramide synthase 3b |
| chr9_-_56459660 | 0.03 |
ENSDART00000162771
|
ubxn4
|
UBX domain protein 4 |
| chr22_-_8594846 | 0.02 |
ENSDART00000081690
ENSDART00000192241 |
si:ch73-27e22.1
|
si:ch73-27e22.1 |
| chr11_-_8269227 | 0.02 |
ENSDART00000127202
ENSDART00000158079 ENSDART00000158748 |
si:cabz01021067.1
pimr202
|
si:cabz01021067.1 Pim proto-oncogene, serine/threonine kinase, related 202 |
| chr9_-_56420087 | 0.01 |
ENSDART00000112016
|
actr3
|
ARP3 actin related protein 3 homolog |
| chr16_-_50918908 | 0.01 |
ENSDART00000108744
|
CU855821.1
|
|
| chr22_-_16412496 | 0.00 |
ENSDART00000137497
|
cts12
|
cathepsin 12 |
Network of associatons between targets according to the STRING database.
First level regulatory network of klf3+klf8
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.6 | 3.9 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.3 | 0.8 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.2 | 0.7 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.2 | 0.9 | GO:0050955 | thermoception(GO:0050955) detection of temperature stimulus involved in thermoception(GO:0050960) detection of temperature stimulus involved in sensory perception(GO:0050961) |
| 0.2 | 0.6 | GO:0071435 | potassium ion export(GO:0071435) potassium ion export across plasma membrane(GO:0097623) |
| 0.1 | 0.5 | GO:0051204 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.1 | 2.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 3.4 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.1 | 0.9 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.8 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 | 0.7 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.3 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.1 | 1.1 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.1 | 0.9 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 1.7 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.2 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.0 | 0.5 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 2.8 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.0 | 0.5 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 3.3 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 1.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.5 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.5 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 2.3 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.4 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.2 | GO:0030819 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 1.0 | GO:0009308 | amine metabolic process(GO:0009308) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 0.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.9 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 4.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 2.1 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.5 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 2.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.8 | GO:0016342 | catenin complex(GO:0016342) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 1.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.6 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 2.5 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.1 | 1.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 1.1 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 3.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.5 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.1 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.5 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.4 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.9 | GO:0015278 | calcium-release channel activity(GO:0015278) |
| 0.0 | 0.5 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 1.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0030955 | potassium ion binding(GO:0030955) alkali metal ion binding(GO:0031420) |
| 0.0 | 4.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 3.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 2.3 | GO:0042626 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 3.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 0.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.0 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 3.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |