Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
Results for mecp2
Z-value: 1.34
Transcription factors associated with mecp2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mecp2
|
ENSDARG00000014218 | methyl CpG binding protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mecp2 | dr11_v1_chr8_-_7637626_7637640 | 0.91 | 6.6e-04 | Click! |
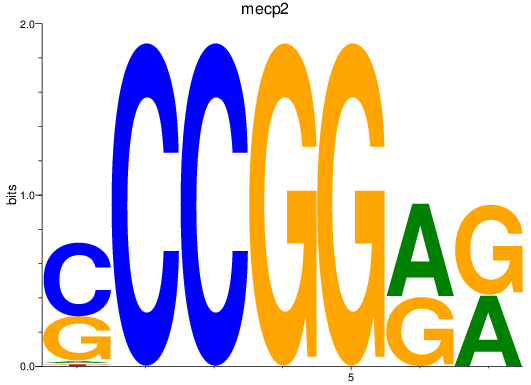
Activity profile of mecp2 motif
Sorted Z-values of mecp2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_55758103 | 1.71 |
ENSDART00000185964
|
CT583728.23
|
|
| chr9_-_32158288 | 1.53 |
ENSDART00000037182
|
ankrd44
|
ankyrin repeat domain 44 |
| chr5_-_825920 | 1.45 |
ENSDART00000126982
|
zgc:158463
|
zgc:158463 |
| chr4_+_3438510 | 1.38 |
ENSDART00000155320
|
atxn7l1
|
ataxin 7-like 1 |
| chr4_-_77561679 | 1.29 |
ENSDART00000180809
|
AL935186.9
|
|
| chr11_+_42422371 | 1.29 |
ENSDART00000163780
|
dennd6aa
|
DENN/MADD domain containing 6Aa |
| chr19_-_47587719 | 1.27 |
ENSDART00000111108
|
CABZ01071972.1
|
|
| chr19_-_27830818 | 1.27 |
ENSDART00000131767
|
papd7
|
PAP associated domain containing 7 |
| chr5_-_30382925 | 1.21 |
ENSDART00000125381
|
gig2o
|
grass carp reovirus (GCRV)-induced gene 2o |
| chr17_-_27235797 | 1.21 |
ENSDART00000130080
|
asap3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| chr8_+_48966165 | 1.14 |
ENSDART00000165425
|
aak1a
|
AP2 associated kinase 1a |
| chr12_+_24060894 | 1.12 |
ENSDART00000021298
|
asb3
|
ankyrin repeat and SOCS box containing 3 |
| chr8_+_48965767 | 1.08 |
ENSDART00000008058
|
aak1a
|
AP2 associated kinase 1a |
| chr23_+_553396 | 1.08 |
ENSDART00000034707
|
lsm14b
|
LSM family member 14B |
| chr20_+_32523576 | 1.01 |
ENSDART00000147319
|
scml4
|
Scm polycomb group protein like 4 |
| chr23_-_32156278 | 1.01 |
ENSDART00000157479
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr7_-_51773166 | 1.00 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr23_+_28128453 | 0.99 |
ENSDART00000182618
|
c1galt1a
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1a |
| chr19_+_41479990 | 0.99 |
ENSDART00000087187
|
ago2
|
argonaute RISC catalytic component 2 |
| chr17_+_52300018 | 0.99 |
ENSDART00000190302
|
esrrb
|
estrogen-related receptor beta |
| chr2_-_38287987 | 0.97 |
ENSDART00000185329
ENSDART00000061677 |
si:ch211-14a17.6
|
si:ch211-14a17.6 |
| chr23_-_44819100 | 0.93 |
ENSDART00000076373
|
st8sia7.1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 7.1 |
| chr18_+_18000695 | 0.93 |
ENSDART00000146898
|
si:ch211-212o1.2
|
si:ch211-212o1.2 |
| chr9_-_32343673 | 0.92 |
ENSDART00000078499
|
rftn2
|
raftlin family member 2 |
| chr11_+_42422638 | 0.90 |
ENSDART00000042599
ENSDART00000181175 |
dennd6aa
|
DENN/MADD domain containing 6Aa |
| chr7_+_16348835 | 0.90 |
ENSDART00000002449
|
mpped2a
|
metallophosphoesterase domain containing 2a |
| chr13_-_44782462 | 0.90 |
ENSDART00000141298
ENSDART00000099990 |
btbd9
|
BTB (POZ) domain containing 9 |
| chr1_-_23308225 | 0.88 |
ENSDART00000137567
ENSDART00000008201 |
smim14
|
small integral membrane protein 14 |
| chr19_-_11846958 | 0.88 |
ENSDART00000148516
|
ctdp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
| chr11_+_39969048 | 0.87 |
ENSDART00000193693
|
per3
|
period circadian clock 3 |
| chr21_+_13233377 | 0.87 |
ENSDART00000142569
|
specc1lb
|
sperm antigen with calponin homology and coiled-coil domains 1-like b |
| chr10_-_9115383 | 0.87 |
ENSDART00000139324
|
si:dkeyp-41f9.3
|
si:dkeyp-41f9.3 |
| chr11_-_25853212 | 0.86 |
ENSDART00000145655
|
tmem51b
|
transmembrane protein 51b |
| chr19_+_14454306 | 0.86 |
ENSDART00000161965
|
zdhhc18b
|
zinc finger, DHHC-type containing 18b |
| chr16_-_42066523 | 0.85 |
ENSDART00000180538
ENSDART00000058620 |
zp3d.1
|
zona pellucida glycoprotein 3d tandem duplicate 1 |
| chr23_+_43668756 | 0.85 |
ENSDART00000112598
|
otud4
|
OTU deubiquitinase 4 |
| chr3_+_35005062 | 0.83 |
ENSDART00000181163
|
prkcbb
|
protein kinase C, beta b |
| chr15_-_47895200 | 0.83 |
ENSDART00000027060
|
DMWD
|
zmp:0000000529 |
| chr15_+_38299563 | 0.82 |
ENSDART00000099375
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr20_-_51547464 | 0.82 |
ENSDART00000099486
|
disp1
|
dispatched homolog 1 (Drosophila) |
| chr15_-_30815826 | 0.81 |
ENSDART00000156160
ENSDART00000145918 |
msi2b
|
musashi RNA-binding protein 2b |
| chr20_-_52939501 | 0.81 |
ENSDART00000166508
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr21_-_2814709 | 0.81 |
ENSDART00000097664
|
SEMA4D
|
semaphorin 4D |
| chr19_+_13994563 | 0.80 |
ENSDART00000164696
|
tmem222b
|
transmembrane protein 222b |
| chr5_-_9216758 | 0.76 |
ENSDART00000134896
ENSDART00000147000 |
lrp13
|
low-density lipoprotein receptor related-protein 13 |
| chr21_-_13751535 | 0.75 |
ENSDART00000111666
|
npdc1a
|
neural proliferation, differentiation and control, 1a |
| chr10_+_39476600 | 0.75 |
ENSDART00000135756
|
kirrel3a
|
kirre like nephrin family adhesion molecule 3a |
| chr8_-_22739757 | 0.73 |
ENSDART00000182167
ENSDART00000171891 |
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr5_+_45140914 | 0.71 |
ENSDART00000172702
|
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr9_-_135774 | 0.70 |
ENSDART00000160435
|
FQ377903.1
|
|
| chr15_+_23208042 | 0.70 |
ENSDART00000006085
|
cbl
|
Cbl proto-oncogene, E3 ubiquitin protein ligase |
| chr20_+_474288 | 0.69 |
ENSDART00000026794
|
nt5dc1
|
5'-nucleotidase domain containing 1 |
| chr6_+_41554794 | 0.69 |
ENSDART00000165424
|
srgap3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr10_+_10972795 | 0.66 |
ENSDART00000127331
|
cdc37l1
|
cell division cycle 37-like 1 |
| chr9_+_17787864 | 0.66 |
ENSDART00000013111
|
dgkh
|
diacylglycerol kinase, eta |
| chr3_+_14463941 | 0.65 |
ENSDART00000170927
|
cnn1b
|
calponin 1, basic, smooth muscle, b |
| chr1_-_55248496 | 0.65 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr2_+_25657958 | 0.64 |
ENSDART00000161407
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr7_+_32021669 | 0.62 |
ENSDART00000173976
|
mettl15
|
methyltransferase like 15 |
| chr2_+_25658112 | 0.61 |
ENSDART00000051234
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr25_+_2776511 | 0.61 |
ENSDART00000115280
|
neo1b
|
neogenin 1b |
| chr23_-_2037566 | 0.61 |
ENSDART00000191312
ENSDART00000127443 |
prdm5
|
PR domain containing 5 |
| chr11_-_27917730 | 0.60 |
ENSDART00000173219
|
eif4g3a
|
eukaryotic translation initiation factor 4 gamma, 3a |
| chr17_+_43629008 | 0.60 |
ENSDART00000184185
ENSDART00000181681 |
znf365
|
zinc finger protein 365 |
| chr16_-_43317927 | 0.60 |
ENSDART00000164472
|
washc5
|
WASH complex subunit 5 |
| chr8_+_50190742 | 0.59 |
ENSDART00000099863
|
slc25a37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr20_-_16078741 | 0.59 |
ENSDART00000021550
|
ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr17_-_20202725 | 0.58 |
ENSDART00000133650
|
ecd
|
ecdysoneless homolog (Drosophila) |
| chr15_+_45595385 | 0.58 |
ENSDART00000161937
ENSDART00000170214 ENSDART00000157450 |
atg16l1
|
ATG16 autophagy related 16-like 1 (S. cerevisiae) |
| chr21_-_30031396 | 0.57 |
ENSDART00000157167
|
pwwp2a
|
PWWP domain containing 2A |
| chr19_-_31576321 | 0.57 |
ENSDART00000103612
|
tdp2b
|
tyrosyl-DNA phosphodiesterase 2b |
| chr17_-_23709347 | 0.57 |
ENSDART00000124661
|
papss2a
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2a |
| chr18_+_18405992 | 0.57 |
ENSDART00000080174
|
n4bp1
|
nedd4 binding protein 1 |
| chr12_-_13730501 | 0.57 |
ENSDART00000152370
|
foxh1
|
forkhead box H1 |
| chr25_+_4581214 | 0.56 |
ENSDART00000185552
|
CABZ01068600.1
|
|
| chr12_-_13966184 | 0.56 |
ENSDART00000066368
|
klhl11
|
kelch-like family member 11 |
| chr8_+_23165749 | 0.56 |
ENSDART00000063057
|
dnajc5aa
|
DnaJ (Hsp40) homolog, subfamily C, member 5aa |
| chr7_+_53254234 | 0.55 |
ENSDART00000169830
|
trip4
|
thyroid hormone receptor interactor 4 |
| chr5_-_24712405 | 0.55 |
ENSDART00000033630
|
si:ch211-106a19.1
|
si:ch211-106a19.1 |
| chr4_-_12102025 | 0.55 |
ENSDART00000048391
ENSDART00000023894 |
braf
|
B-Raf proto-oncogene, serine/threonine kinase |
| chr5_+_15992655 | 0.55 |
ENSDART00000182148
|
znrf3
|
zinc and ring finger 3 |
| chr18_+_50961953 | 0.54 |
ENSDART00000158768
|
ppfia1
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 |
| chr3_-_10582384 | 0.54 |
ENSDART00000048095
ENSDART00000155152 |
elac2
|
elaC ribonuclease Z 2 |
| chr11_+_2687395 | 0.54 |
ENSDART00000082510
|
b3galt6
|
UDP-Gal:betaGal beta 1,3-galactosyltransferase polypeptide 6 |
| chr3_+_48842918 | 0.53 |
ENSDART00000159420
|
hs3st3b1a
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1a |
| chr4_+_23223881 | 0.53 |
ENSDART00000133056
ENSDART00000089126 |
trhde.1
|
thyrotropin releasing hormone degrading enzyme, tandem duplicate 1 |
| chr18_-_399554 | 0.53 |
ENSDART00000164374
ENSDART00000186311 ENSDART00000181816 ENSDART00000181892 |
si:ch211-79l17.1
|
si:ch211-79l17.1 |
| chr17_+_43926523 | 0.53 |
ENSDART00000121550
ENSDART00000041447 |
ktn1
|
kinectin 1 |
| chr7_-_33829824 | 0.53 |
ENSDART00000074729
|
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr8_+_14915332 | 0.52 |
ENSDART00000164385
|
abl2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr8_+_18555559 | 0.52 |
ENSDART00000149523
|
tab3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr11_-_45171139 | 0.52 |
ENSDART00000167036
ENSDART00000161712 ENSDART00000158156 |
syngr2b
|
synaptogyrin 2b |
| chr10_+_31809226 | 0.52 |
ENSDART00000087898
|
foxo1b
|
forkhead box O1 b |
| chr20_+_34502606 | 0.52 |
ENSDART00000139739
|
gorab
|
golgin, rab6-interacting |
| chr24_+_24285751 | 0.51 |
ENSDART00000122294
|
ythdf3
|
YTH N(6)-methyladenosine RNA binding protein 3 |
| chr14_+_35024521 | 0.50 |
ENSDART00000158634
ENSDART00000170631 |
ebf3a
|
early B cell factor 3a |
| chr16_-_13388821 | 0.50 |
ENSDART00000144062
|
grin2db
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, b |
| chr11_-_20987378 | 0.50 |
ENSDART00000110140
|
taf4a
|
TAF4A RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr14_-_27289042 | 0.49 |
ENSDART00000159727
|
pcdh11
|
protocadherin 11 |
| chr15_-_34558579 | 0.49 |
ENSDART00000140882
|
ispd
|
isoprenoid synthase domain containing |
| chr3_-_27880229 | 0.49 |
ENSDART00000151404
|
abat
|
4-aminobutyrate aminotransferase |
| chr12_+_4220353 | 0.49 |
ENSDART00000133675
|
mapk7
|
mitogen-activated protein kinase 7 |
| chr10_-_33156789 | 0.49 |
ENSDART00000192268
ENSDART00000182065 ENSDART00000081170 |
cux1a
|
cut-like homeobox 1a |
| chr15_-_25367309 | 0.48 |
ENSDART00000047471
|
cluha
|
clustered mitochondria (cluA/CLU1) homolog a |
| chr23_+_12361899 | 0.48 |
ENSDART00000143728
|
pigt
|
phosphatidylinositol glycan anchor biosynthesis, class T |
| chr8_+_47897734 | 0.47 |
ENSDART00000140266
|
mfn2
|
mitofusin 2 |
| chr10_-_32494304 | 0.47 |
ENSDART00000028161
|
uvrag
|
UV radiation resistance associated gene |
| chr21_-_14692119 | 0.47 |
ENSDART00000123047
|
ehmt1b
|
euchromatic histone-lysine N-methyltransferase 1b |
| chr2_+_23222939 | 0.47 |
ENSDART00000026800
|
kifap3b
|
kinesin-associated protein 3b |
| chr2_+_54327160 | 0.47 |
ENSDART00000182320
|
LO018508.1
|
|
| chr2_+_39108339 | 0.46 |
ENSDART00000085675
|
clstn2
|
calsyntenin 2 |
| chr19_+_232536 | 0.46 |
ENSDART00000137880
|
syngap1a
|
synaptic Ras GTPase activating protein 1a |
| chr19_+_32979331 | 0.46 |
ENSDART00000078066
|
spire1a
|
spire-type actin nucleation factor 1a |
| chr25_+_33972019 | 0.45 |
ENSDART00000188515
ENSDART00000121498 |
ice2
|
interactor of little elongator complex ELL subunit 2 |
| chr1_+_15258641 | 0.45 |
ENSDART00000033018
|
pi4k2b
|
phosphatidylinositol 4-kinase type 2 beta |
| chr4_-_12930086 | 0.45 |
ENSDART00000013604
|
lemd3
|
LEM domain containing 3 |
| chr7_+_1442059 | 0.45 |
ENSDART00000173391
|
si:cabz01090193.1
|
si:cabz01090193.1 |
| chr14_+_25505468 | 0.45 |
ENSDART00000079016
|
thoc3
|
THO complex 3 |
| chr21_-_25613249 | 0.44 |
ENSDART00000137896
|
fibpb
|
fibroblast growth factor (acidic) intracellular binding protein b |
| chr5_+_69950882 | 0.44 |
ENSDART00000097359
|
dnajc25
|
DnaJ (Hsp40) homolog, subfamily C , member 25 |
| chr8_+_1843135 | 0.44 |
ENSDART00000141452
|
snap29
|
synaptosomal-associated protein 29 |
| chr6_-_36795111 | 0.43 |
ENSDART00000160669
ENSDART00000104256 ENSDART00000187751 ENSDART00000161928 ENSDART00000183264 |
opa1
|
optic atrophy 1 (autosomal dominant) |
| chr21_-_11367271 | 0.43 |
ENSDART00000151000
ENSDART00000151465 |
zgc:162472
|
zgc:162472 |
| chr21_+_10021823 | 0.43 |
ENSDART00000163995
|
herc7
|
hect domain and RLD 7 |
| chr15_+_45994123 | 0.43 |
ENSDART00000124704
|
lrfn1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr14_-_6402769 | 0.43 |
ENSDART00000121552
|
slc44a1b
|
solute carrier family 44 (choline transporter), member 1b |
| chr20_+_46699021 | 0.43 |
ENSDART00000167398
ENSDART00000029894 |
wdr26b
|
WD repeat domain 26b |
| chr3_+_20001608 | 0.43 |
ENSDART00000137944
|
asb16
|
ankyrin repeat and SOCS box containing 16 |
| chr8_+_2757821 | 0.43 |
ENSDART00000051403
ENSDART00000160551 |
sh3glb2a
|
SH3-domain GRB2-like endophilin B2a |
| chr23_+_20431140 | 0.43 |
ENSDART00000193950
|
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr13_-_36525982 | 0.42 |
ENSDART00000114744
|
pacs2
|
phosphofurin acidic cluster sorting protein 2 |
| chr19_-_9503473 | 0.42 |
ENSDART00000091615
|
iffo1a
|
intermediate filament family orphan 1a |
| chr25_-_12203952 | 0.42 |
ENSDART00000158204
ENSDART00000091727 |
ntrk3a
|
neurotrophic tyrosine kinase, receptor, type 3a |
| chr15_-_34871968 | 0.42 |
ENSDART00000191980
|
mgat1a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase a |
| chr15_+_44093286 | 0.42 |
ENSDART00000114352
|
zgc:112998
|
zgc:112998 |
| chr16_+_42471455 | 0.41 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr1_+_52632856 | 0.41 |
ENSDART00000011725
|
slc44a1a
|
solute carrier family 44 (choline transporter), member 1a |
| chr2_-_9744081 | 0.41 |
ENSDART00000097732
|
dvl3a
|
dishevelled segment polarity protein 3a |
| chr12_-_33789006 | 0.41 |
ENSDART00000034550
|
llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr16_-_2807527 | 0.41 |
ENSDART00000148927
|
tgfb2
|
transforming growth factor, beta 2 |
| chr20_+_30702531 | 0.41 |
ENSDART00000062525
|
acbd3
|
acyl-Coenzyme A binding domain containing 3 |
| chr10_-_6588793 | 0.40 |
ENSDART00000163788
|
chd1
|
chromodomain helicase DNA binding protein 1 |
| chr3_+_22059066 | 0.40 |
ENSDART00000155739
|
kansl1b
|
KAT8 regulatory NSL complex subunit 1b |
| chr18_-_21910991 | 0.40 |
ENSDART00000089787
ENSDART00000169220 ENSDART00000132381 ENSDART00000191764 |
edc4
|
enhancer of mRNA decapping 4 |
| chr22_-_4398069 | 0.40 |
ENSDART00000181893
|
kdm4b
|
lysine (K)-specific demethylase 4B |
| chr12_-_18961289 | 0.40 |
ENSDART00000168405
|
ep300a
|
E1A binding protein p300 a |
| chr3_-_13461056 | 0.39 |
ENSDART00000137678
|
fbxw9
|
F-box and WD repeat domain containing 9 |
| chr3_+_17653784 | 0.39 |
ENSDART00000159984
ENSDART00000157682 ENSDART00000187937 |
kat2a
|
K(lysine) acetyltransferase 2A |
| chr22_+_1006573 | 0.39 |
ENSDART00000123458
|
pparda
|
peroxisome proliferator-activated receptor delta a |
| chr11_-_21363834 | 0.39 |
ENSDART00000080051
|
RASSF5
|
si:dkey-85p17.3 |
| chr12_+_33038757 | 0.39 |
ENSDART00000153146
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr13_+_14006118 | 0.39 |
ENSDART00000131875
ENSDART00000089528 |
atrn
|
attractin |
| chr13_-_36566260 | 0.38 |
ENSDART00000030133
|
synj2bp
|
synaptojanin 2 binding protein |
| chr23_+_10426219 | 0.38 |
ENSDART00000139100
|
eif4ba
|
eukaryotic translation initiation factor 4Ba |
| chr13_-_36911118 | 0.38 |
ENSDART00000048739
|
trim9
|
tripartite motif containing 9 |
| chr15_+_30158652 | 0.37 |
ENSDART00000190682
|
nlk2
|
nemo-like kinase, type 2 |
| chr7_+_1534820 | 0.37 |
ENSDART00000192997
|
tox4b
|
TOX high mobility group box family member 4 b |
| chr11_+_41459408 | 0.37 |
ENSDART00000182285
|
park7
|
parkinson protein 7 |
| chr23_+_20431388 | 0.37 |
ENSDART00000132920
ENSDART00000102963 ENSDART00000109899 ENSDART00000140219 |
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr18_-_33344 | 0.37 |
ENSDART00000129125
|
pde8a
|
phosphodiesterase 8A |
| chr9_+_1505206 | 0.36 |
ENSDART00000093427
ENSDART00000137230 |
pde11a
|
phosphodiesterase 11a |
| chr12_+_36428052 | 0.36 |
ENSDART00000131300
|
unk
|
unkempt family zinc finger |
| chr10_-_41156348 | 0.36 |
ENSDART00000058622
|
aak1b
|
AP2 associated kinase 1b |
| chr9_-_52206336 | 0.36 |
ENSDART00000114222
|
tanc1b
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1b |
| chr7_+_41887429 | 0.36 |
ENSDART00000115090
|
gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr16_+_33953644 | 0.36 |
ENSDART00000164447
ENSDART00000159969 |
arid1aa
|
AT rich interactive domain 1Aa (SWI-like) |
| chr15_+_39096736 | 0.36 |
ENSDART00000129511
ENSDART00000014877 |
robo2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
| chr1_+_40613297 | 0.35 |
ENSDART00000040798
ENSDART00000168067 ENSDART00000130490 |
naa15b
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit b |
| chr20_-_3276339 | 0.35 |
ENSDART00000166831
|
rps6kc1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr19_-_7441686 | 0.35 |
ENSDART00000168194
|
gabpb2a
|
GA binding protein transcription factor, beta subunit 2a |
| chr14_-_38946808 | 0.35 |
ENSDART00000139293
|
gla
|
galactosidase, alpha |
| chr13_-_24745288 | 0.34 |
ENSDART00000031564
|
sfr1
|
SWI5-dependent homologous recombination repair protein 1 |
| chr10_+_43037064 | 0.33 |
ENSDART00000160159
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr14_+_46118834 | 0.33 |
ENSDART00000124417
ENSDART00000017785 |
naa15a
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit a |
| chr3_-_13068189 | 0.33 |
ENSDART00000167180
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr17_-_50010121 | 0.33 |
ENSDART00000122747
|
tmem30aa
|
transmembrane protein 30Aa |
| chr15_-_45110011 | 0.33 |
ENSDART00000182047
ENSDART00000188662 |
CABZ01072607.1
|
|
| chr12_-_33789218 | 0.33 |
ENSDART00000193258
|
llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr16_+_32136550 | 0.33 |
ENSDART00000147526
|
sphk2
|
sphingosine kinase 2 |
| chr2_-_37401600 | 0.33 |
ENSDART00000015723
|
prkci
|
protein kinase C, iota |
| chr13_+_11876437 | 0.33 |
ENSDART00000179753
|
trim8a
|
tripartite motif containing 8a |
| chr5_+_4016271 | 0.32 |
ENSDART00000113627
ENSDART00000105832 ENSDART00000121415 |
ggnbp2
|
gametogenetin binding protein 2 |
| chr16_+_27345383 | 0.32 |
ENSDART00000078250
ENSDART00000162857 |
nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr16_-_12914288 | 0.32 |
ENSDART00000184221
|
cacng8b
|
calcium channel, voltage-dependent, gamma subunit 8b |
| chr8_-_11131695 | 0.32 |
ENSDART00000055742
|
nras
|
NRAS proto-oncogene, GTPase |
| chr3_-_25054002 | 0.32 |
ENSDART00000086768
|
ep300b
|
E1A binding protein p300 b |
| chr2_+_51645164 | 0.32 |
ENSDART00000169600
|
abhd4
|
abhydrolase domain containing 4 |
| chr21_-_30030644 | 0.32 |
ENSDART00000190810
|
CU855895.2
|
|
| chr19_+_47301893 | 0.32 |
ENSDART00000051695
|
tpmt.1
|
thiopurine S-methyltransferase, tandem duplicate 1 |
| chr5_-_22573624 | 0.32 |
ENSDART00000131889
ENSDART00000080886 ENSDART00000147513 ENSDART00000080882 |
aifm1
|
apoptosis-inducing factor, mitochondrion-associated 1 |
| chr18_-_46369516 | 0.32 |
ENSDART00000018163
|
irf2bp1
|
interferon regulatory factor 2 binding protein 1 |
| chr24_+_5208171 | 0.32 |
ENSDART00000155926
ENSDART00000154464 |
si:ch73-206p6.1
|
si:ch73-206p6.1 |
| chr7_+_5906327 | 0.32 |
ENSDART00000173160
|
zgc:112234
|
zgc:112234 |
| chr6_+_33931740 | 0.31 |
ENSDART00000130492
ENSDART00000151213 |
orc1
|
origin recognition complex, subunit 1 |
| chr2_-_43739740 | 0.31 |
ENSDART00000113849
|
kif5ba
|
kinesin family member 5B, a |
| chr19_+_3056450 | 0.31 |
ENSDART00000141324
ENSDART00000082353 |
hsf1
|
heat shock transcription factor 1 |
| chr7_+_32021982 | 0.31 |
ENSDART00000173848
|
mettl15
|
methyltransferase like 15 |
| chr7_-_73851280 | 0.31 |
ENSDART00000190053
|
FP236812.3
|
|
| chr6_-_59357256 | 0.30 |
ENSDART00000074534
|
fam210b
|
family with sequence similarity 210, member B |
| chr3_+_1211242 | 0.30 |
ENSDART00000171287
ENSDART00000165769 |
poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of mecp2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.3 | 0.9 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.3 | 0.9 | GO:0097095 | frontonasal suture morphogenesis(GO:0097095) |
| 0.3 | 1.0 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.2 | 1.0 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.2 | 0.8 | GO:2000660 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.2 | 1.0 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.2 | 0.6 | GO:0034036 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.2 | 0.6 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.2 | 0.7 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.2 | 0.5 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.2 | 0.2 | GO:0007618 | mating(GO:0007618) |
| 0.2 | 0.6 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.2 | 2.6 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.2 | 0.5 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 0.4 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.1 | 1.3 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.7 | GO:0010719 | photoreceptor cell morphogenesis(GO:0008594) negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.1 | 0.5 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.1 | 0.5 | GO:0042779 | tRNA 3'-trailer cleavage(GO:0042779) |
| 0.1 | 0.5 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.1 | 0.4 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.1 | 0.7 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.6 | GO:0039694 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 1.8 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.8 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.1 | 0.5 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.1 | 0.7 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.9 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 0.3 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.6 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 0.3 | GO:0072388 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.1 | 0.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.4 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.1 | 0.2 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 | 0.5 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 0.5 | GO:0048660 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.1 | 0.4 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.3 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.3 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 0.4 | GO:0090179 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.8 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.1 | 0.5 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.1 | 0.2 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.1 | 0.4 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.1 | 0.2 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.1 | 0.4 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 1.0 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.3 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.0 | 0.5 | GO:0050779 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.0 | 0.6 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.4 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.3 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.4 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.6 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.9 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.3 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.4 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.9 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.5 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.3 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.0 | 0.6 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.7 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.6 | GO:0051567 | histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 0.0 | 0.1 | GO:0071869 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.0 | 0.3 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.5 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.4 | GO:1903010 | regulation of bone development(GO:1903010) |
| 0.0 | 0.1 | GO:1905132 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.0 | 0.2 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.0 | 0.4 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.6 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 | 0.7 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.6 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.2 | GO:0089700 | protein kinase D signaling(GO:0089700) regulation of lymphangiogenesis(GO:1901490) |
| 0.0 | 0.7 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.3 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 1.4 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.6 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.3 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 0.4 | GO:1990090 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.0 | 0.1 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.2 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.3 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 0.2 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.6 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.5 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.1 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.2 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 0.0 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.7 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.1 | GO:0032988 | ribosome disassembly(GO:0032790) ribonucleoprotein complex disassembly(GO:0032988) |
| 0.0 | 0.4 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.3 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.3 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 1.4 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.0 | 0.2 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:0001765 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.4 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.7 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 1.0 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.0 | 0.6 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.1 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.0 | GO:0042416 | dopamine biosynthetic process from tyrosine(GO:0006585) dopamine biosynthetic process(GO:0042416) |
| 0.0 | 0.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.3 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.4 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.3 | GO:0043113 | receptor clustering(GO:0043113) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.1 | 0.7 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.6 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.4 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.1 | 0.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.6 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.1 | 0.4 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.2 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.1 | 0.4 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 0.6 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.4 | GO:0035060 | brahma complex(GO:0035060) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.2 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.9 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.2 | GO:0035032 | extrinsic component of vacuolar membrane(GO:0000306) phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.5 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 0.2 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 1.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.4 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.3 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.7 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.5 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 2.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.8 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.4 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.6 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.1 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.2 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.5 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 1.0 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 1.1 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.4 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.3 | 0.8 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.2 | 1.0 | GO:0048531 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.2 | 1.0 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.2 | 0.7 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.2 | 2.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 0.6 | GO:0030882 | lipid antigen binding(GO:0030882) |
| 0.2 | 0.6 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.2 | 0.6 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.2 | 0.5 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.1 | 0.4 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 1.0 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 1.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.4 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 0.8 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 0.9 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.3 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.1 | 0.3 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.1 | 0.3 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.9 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 1.8 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.3 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.1 | 0.4 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 0.4 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.9 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.3 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.5 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.3 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.1 | 0.4 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 0.4 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.4 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.6 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 0.5 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.2 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.1 | 0.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.2 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.1 | 0.3 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.4 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.1 | 0.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.8 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 1.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.5 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.7 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.3 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.7 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 1.4 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.7 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.5 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.0 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.6 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.8 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.5 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.7 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.7 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.1 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.2 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.7 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.8 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.3 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.2 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 0.7 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 0.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.3 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.9 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 1.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.5 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.5 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |