Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
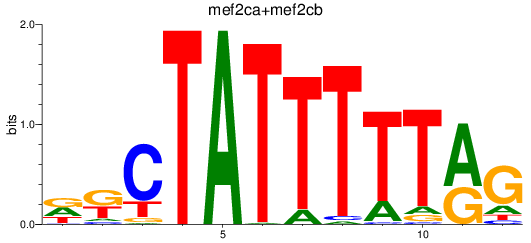
Results for mef2ca+mef2cb
Z-value: 1.09
Transcription factors associated with mef2ca+mef2cb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mef2cb
|
ENSDARG00000009418 | myocyte enhancer factor 2cb |
|
mef2ca
|
ENSDARG00000029764 | myocyte enhancer factor 2ca |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mef2ca | dr11_v1_chr10_-_43568239_43568241 | 0.89 | 1.3e-03 | Click! |
| mef2cb | dr11_v1_chr5_-_48260145_48260209 | 0.80 | 9.5e-03 | Click! |
Activity profile of mef2ca+mef2cb motif
Sorted Z-values of mef2ca+mef2cb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_61205711 | 2.23 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr15_-_23645810 | 1.69 |
ENSDART00000168845
|
ckmb
|
creatine kinase, muscle b |
| chr9_-_33107237 | 1.66 |
ENSDART00000013918
|
casq2
|
calsequestrin 2 |
| chr1_+_7546259 | 1.61 |
ENSDART00000015732
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr25_+_29160102 | 1.55 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr11_+_11201096 | 1.55 |
ENSDART00000171916
ENSDART00000171521 ENSDART00000087105 ENSDART00000159603 |
myom2a
|
myomesin 2a |
| chr25_+_20089986 | 1.47 |
ENSDART00000143441
ENSDART00000184073 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr5_-_72125551 | 1.35 |
ENSDART00000149412
|
smyd1a
|
SET and MYND domain containing 1a |
| chr16_+_29663809 | 1.29 |
ENSDART00000191336
|
tmod4
|
tropomodulin 4 (muscle) |
| chr6_+_3680651 | 1.23 |
ENSDART00000013588
|
klhl41b
|
kelch-like family member 41b |
| chr23_-_27633730 | 1.22 |
ENSDART00000103639
|
arf3a
|
ADP-ribosylation factor 3a |
| chr3_+_28953274 | 1.20 |
ENSDART00000133528
ENSDART00000103602 |
lgals2a
|
lectin, galactoside-binding, soluble, 2a |
| chr4_-_9592402 | 1.19 |
ENSDART00000114060
|
cdnf
|
cerebral dopamine neurotrophic factor |
| chr12_-_26064480 | 1.19 |
ENSDART00000158215
ENSDART00000171206 ENSDART00000171212 ENSDART00000182956 ENSDART00000186779 |
ldb3b
|
LIM domain binding 3b |
| chr9_-_7673856 | 1.15 |
ENSDART00000102715
|
tuba8l3
|
tubulin, alpha 8 like 3 |
| chr12_-_26064105 | 1.15 |
ENSDART00000168825
|
ldb3b
|
LIM domain binding 3b |
| chr11_+_11200550 | 1.10 |
ENSDART00000181339
ENSDART00000187116 |
myom2a
|
myomesin 2a |
| chr12_+_18524953 | 1.08 |
ENSDART00000090332
|
neurl2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr5_+_51597677 | 1.08 |
ENSDART00000048210
ENSDART00000184797 |
ckmt2b
|
creatine kinase, mitochondrial 2b (sarcomeric) |
| chr12_-_17707449 | 1.05 |
ENSDART00000142427
ENSDART00000034914 |
pvalb3
|
parvalbumin 3 |
| chr25_-_31396479 | 1.04 |
ENSDART00000156828
|
prr33
|
proline rich 33 |
| chr5_+_67390645 | 1.03 |
ENSDART00000014822
|
ebf2
|
early B cell factor 2 |
| chr10_-_8032885 | 1.02 |
ENSDART00000188619
|
atp6v0a2a
|
ATPase H+ transporting V0 subunit a2a |
| chr2_+_30916188 | 0.96 |
ENSDART00000137012
|
myom1a
|
myomesin 1a (skelemin) |
| chr16_+_34523515 | 0.94 |
ENSDART00000041007
|
stmn1b
|
stathmin 1b |
| chr15_-_9031996 | 0.94 |
ENSDART00000124998
|
rtn2a
|
reticulon 2a |
| chr24_-_40700596 | 0.93 |
ENSDART00000162635
|
smyhc2
|
slow myosin heavy chain 2 |
| chr14_-_30366196 | 0.92 |
ENSDART00000007022
|
pdgfrl
|
platelet-derived growth factor receptor-like |
| chr9_-_42873700 | 0.91 |
ENSDART00000125953
|
ttn.1
|
titin, tandem duplicate 1 |
| chr10_-_22803740 | 0.90 |
ENSDART00000079469
ENSDART00000187968 ENSDART00000122543 |
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr10_-_8033468 | 0.87 |
ENSDART00000140476
|
atp6v0a2a
|
ATPase H+ transporting V0 subunit a2a |
| chr11_-_18253111 | 0.87 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr10_+_32104305 | 0.85 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr20_-_9980318 | 0.84 |
ENSDART00000080664
|
ACTC1
|
zgc:86709 |
| chr17_-_5583345 | 0.84 |
ENSDART00000035944
|
clic5a
|
chloride intracellular channel 5a |
| chr12_+_6002715 | 0.84 |
ENSDART00000114961
|
si:ch211-131k2.3
|
si:ch211-131k2.3 |
| chr5_+_67390115 | 0.83 |
ENSDART00000193255
|
ebf2
|
early B cell factor 2 |
| chr8_-_27687095 | 0.82 |
ENSDART00000086946
|
mov10b.1
|
Moloney leukemia virus 10b, tandem duplicate 1 |
| chr9_-_6927587 | 0.81 |
ENSDART00000059092
|
tmem182a
|
transmembrane protein 182a |
| chr4_+_21717793 | 0.79 |
ENSDART00000040266
|
myf6
|
myogenic factor 6 |
| chr17_-_12336987 | 0.78 |
ENSDART00000172001
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr7_-_13381129 | 0.78 |
ENSDART00000164326
|
si:ch73-119p20.1
|
si:ch73-119p20.1 |
| chr10_-_22845485 | 0.77 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr8_+_7322465 | 0.77 |
ENSDART00000192380
|
ssuh2rs1
|
ssu-2 homolog, related sequence 1 |
| chr13_+_22249636 | 0.76 |
ENSDART00000108472
ENSDART00000173123 |
synpo2la
|
synaptopodin 2-like a |
| chr13_+_27951688 | 0.75 |
ENSDART00000050303
|
b3gat2
|
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) |
| chr15_+_40008370 | 0.74 |
ENSDART00000063783
|
itm2ca
|
integral membrane protein 2Ca |
| chr14_-_11430566 | 0.73 |
ENSDART00000137154
ENSDART00000091158 |
irg1l
|
immunoresponsive gene 1, like |
| chr7_-_23745984 | 0.70 |
ENSDART00000048050
|
ITGB1BP2
|
zgc:92429 |
| chr21_-_22730832 | 0.70 |
ENSDART00000101797
|
fbxo40.1
|
F-box protein 40, tandem duplicate 1 |
| chr19_-_41069573 | 0.69 |
ENSDART00000111982
ENSDART00000193142 |
sgce
|
sarcoglycan, epsilon |
| chr9_-_14108896 | 0.68 |
ENSDART00000135209
|
prkag3b
|
protein kinase, AMP-activated, gamma 3b non-catalytic subunit |
| chr5_-_19394440 | 0.68 |
ENSDART00000163771
|
foxn4
|
forkhead box N4 |
| chr6_-_14139503 | 0.66 |
ENSDART00000089577
|
cacnb4b
|
calcium channel, voltage-dependent, beta 4b subunit |
| chr19_-_24443394 | 0.66 |
ENSDART00000090825
|
thbs3b
|
thrombospondin 3b |
| chr11_-_39202915 | 0.64 |
ENSDART00000105133
|
wnt4a
|
wingless-type MMTV integration site family, member 4a |
| chr5_-_48268049 | 0.63 |
ENSDART00000187454
|
mef2cb
|
myocyte enhancer factor 2cb |
| chr23_+_24931999 | 0.62 |
ENSDART00000136162
ENSDART00000140335 |
klhl21
|
kelch-like family member 21 |
| chr10_+_22724059 | 0.61 |
ENSDART00000136123
|
kdm6bb
|
lysine (K)-specific demethylase 6B, b |
| chr16_-_43026273 | 0.61 |
ENSDART00000156820
ENSDART00000189080 |
si:dkey-7j14.5
|
si:dkey-7j14.5 |
| chr8_+_22930627 | 0.61 |
ENSDART00000187860
|
sypa
|
synaptophysin a |
| chr6_+_29410986 | 0.60 |
ENSDART00000065293
|
usp13
|
ubiquitin specific peptidase 13 |
| chr8_+_34731982 | 0.60 |
ENSDART00000066050
|
hpdb
|
4-hydroxyphenylpyruvate dioxygenase b |
| chr20_+_23173710 | 0.60 |
ENSDART00000074172
|
sgcb
|
sarcoglycan, beta (dystrophin-associated glycoprotein) |
| chr19_-_24443867 | 0.60 |
ENSDART00000163763
ENSDART00000043133 |
thbs3b
|
thrombospondin 3b |
| chr7_+_19495905 | 0.59 |
ENSDART00000125584
ENSDART00000173774 |
si:ch211-212k18.8
|
si:ch211-212k18.8 |
| chr8_-_32497815 | 0.59 |
ENSDART00000122359
|
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr25_-_26736088 | 0.59 |
ENSDART00000067114
|
fbxl22
|
F-box and leucine-rich repeat protein 22 |
| chr1_+_50968908 | 0.59 |
ENSDART00000150353
ENSDART00000012842 |
mdh1aa
|
malate dehydrogenase 1Aa, NAD (soluble) |
| chr19_-_38611814 | 0.59 |
ENSDART00000151958
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr23_-_5685023 | 0.59 |
ENSDART00000148680
ENSDART00000149365 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr21_+_19070921 | 0.58 |
ENSDART00000029874
|
nkx6.1
|
NK6 homeobox 1 |
| chr9_-_34300707 | 0.58 |
ENSDART00000049805
|
ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr16_+_20895904 | 0.57 |
ENSDART00000052662
|
hoxa13b
|
homeobox A13b |
| chr4_+_19534833 | 0.56 |
ENSDART00000140028
|
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr5_+_64368770 | 0.56 |
ENSDART00000162246
|
plpp7
|
phospholipid phosphatase 7 (inactive) |
| chr10_-_24371312 | 0.56 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr25_+_5039050 | 0.56 |
ENSDART00000154700
|
parvb
|
parvin, beta |
| chr24_+_20960216 | 0.55 |
ENSDART00000133008
|
si:ch211-161h7.8
|
si:ch211-161h7.8 |
| chr18_+_20494413 | 0.53 |
ENSDART00000060295
|
rapsn
|
receptor-associated protein of the synapse, 43kD |
| chr24_+_20575259 | 0.53 |
ENSDART00000010488
|
klhl40b
|
kelch-like family member 40b |
| chr14_-_21063977 | 0.52 |
ENSDART00000164373
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr1_+_10683843 | 0.52 |
ENSDART00000054879
|
zgc:103678
|
zgc:103678 |
| chr23_+_18722715 | 0.52 |
ENSDART00000137438
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr18_-_15467446 | 0.52 |
ENSDART00000187847
|
endouc
|
endonuclease, polyU-specific C |
| chr7_-_27686021 | 0.51 |
ENSDART00000079112
ENSDART00000100989 |
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr22_+_24715282 | 0.50 |
ENSDART00000088027
ENSDART00000189054 ENSDART00000140430 |
ssx2ipb
|
synovial sarcoma, X breakpoint 2 interacting protein b |
| chr13_+_22480857 | 0.50 |
ENSDART00000078721
ENSDART00000044719 ENSDART00000130957 ENSDART00000078757 ENSDART00000130424 ENSDART00000078747 |
ldb3a
|
LIM domain binding 3a |
| chr9_+_31795343 | 0.50 |
ENSDART00000139584
|
itgbl1
|
integrin, beta-like 1 |
| chr21_-_22951604 | 0.50 |
ENSDART00000083449
ENSDART00000180129 |
dub
|
duboraya |
| chr6_-_49063085 | 0.49 |
ENSDART00000156124
|
si:ch211-105j21.9
|
si:ch211-105j21.9 |
| chr8_+_10304981 | 0.49 |
ENSDART00000160766
|
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr18_+_19974289 | 0.49 |
ENSDART00000090334
ENSDART00000192982 |
skor1b
|
SKI family transcriptional corepressor 1b |
| chr10_+_37145007 | 0.48 |
ENSDART00000131777
|
cuedc1a
|
CUE domain containing 1a |
| chr14_-_12307522 | 0.48 |
ENSDART00000163900
|
myot
|
myotilin |
| chr5_-_36948586 | 0.48 |
ENSDART00000193606
|
h3f3c
|
H3 histone, family 3C |
| chr8_-_32497581 | 0.47 |
ENSDART00000176298
ENSDART00000183340 |
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr9_-_12424791 | 0.47 |
ENSDART00000135447
ENSDART00000088199 |
zgc:162707
|
zgc:162707 |
| chr8_-_18535822 | 0.47 |
ENSDART00000100558
|
nexn
|
nexilin (F actin binding protein) |
| chr8_+_21146262 | 0.47 |
ENSDART00000045684
|
porcn
|
porcupine O-acyltransferase |
| chr20_-_39271844 | 0.47 |
ENSDART00000192708
|
clu
|
clusterin |
| chr20_+_15552657 | 0.46 |
ENSDART00000063912
|
jun
|
Jun proto-oncogene, AP-1 transcription factor subunit |
| chr8_+_52637507 | 0.46 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr9_+_6578580 | 0.46 |
ENSDART00000061577
|
fhl2a
|
four and a half LIM domains 2a |
| chr25_+_16945348 | 0.46 |
ENSDART00000016591
|
fgf6a
|
fibroblast growth factor 6a |
| chr20_+_34455645 | 0.45 |
ENSDART00000135789
|
mettl11b
|
methyltransferase like 11B |
| chr13_-_11536951 | 0.44 |
ENSDART00000018155
|
adss
|
adenylosuccinate synthase |
| chr7_+_27691647 | 0.44 |
ENSDART00000079091
|
cyp2r1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr16_+_26777473 | 0.44 |
ENSDART00000188870
|
cdh17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr13_-_28674422 | 0.44 |
ENSDART00000122754
ENSDART00000057574 |
nt5c2a
|
5'-nucleotidase, cytosolic IIa |
| chr23_+_18722915 | 0.43 |
ENSDART00000025057
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr23_+_24926407 | 0.43 |
ENSDART00000137486
|
klhl21
|
kelch-like family member 21 |
| chr12_+_17106117 | 0.43 |
ENSDART00000149990
|
acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr7_-_33961551 | 0.43 |
ENSDART00000100104
|
skor1a
|
SKI family transcriptional corepressor 1a |
| chr3_-_50046004 | 0.43 |
ENSDART00000109544
|
si:ch1073-100f3.2
|
si:ch1073-100f3.2 |
| chr7_+_41295974 | 0.42 |
ENSDART00000173568
ENSDART00000173544 |
si:dkey-86l18.10
|
si:dkey-86l18.10 |
| chr3_+_59051503 | 0.42 |
ENSDART00000160767
|
rasd4
|
rasd family member 4 |
| chr20_-_26066020 | 0.42 |
ENSDART00000078559
|
myct1a
|
myc target 1a |
| chr3_-_30123113 | 0.42 |
ENSDART00000153562
|
ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr7_+_22543963 | 0.41 |
ENSDART00000101528
|
chrnb1
|
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr3_+_59851537 | 0.41 |
ENSDART00000180997
|
CU693479.1
|
|
| chr16_-_7228276 | 0.41 |
ENSDART00000149030
|
nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr9_-_43142636 | 0.40 |
ENSDART00000134349
ENSDART00000181835 |
ccdc141
|
coiled-coil domain containing 141 |
| chr14_-_21064199 | 0.40 |
ENSDART00000172099
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr15_+_32867420 | 0.40 |
ENSDART00000159442
|
dclk1b
|
doublecortin-like kinase 1b |
| chr22_+_17205608 | 0.40 |
ENSDART00000181267
|
rab3b
|
RAB3B, member RAS oncogene family |
| chr14_+_11430796 | 0.40 |
ENSDART00000165275
|
si:ch211-153b23.3
|
si:ch211-153b23.3 |
| chr1_-_8101495 | 0.40 |
ENSDART00000161938
|
si:dkeyp-9d4.3
|
si:dkeyp-9d4.3 |
| chr2_-_23349116 | 0.39 |
ENSDART00000099690
|
fam129ab
|
family with sequence similarity 129, member Ab |
| chr21_+_12010505 | 0.39 |
ENSDART00000123522
|
aqp7
|
aquaporin 7 |
| chr13_+_13578552 | 0.39 |
ENSDART00000101673
|
foxi2
|
forkhead box I2 |
| chr2_+_20472150 | 0.39 |
ENSDART00000168537
|
agla
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase a |
| chr15_+_43906043 | 0.38 |
ENSDART00000010881
|
naalad2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr19_+_1878500 | 0.38 |
ENSDART00000113392
|
ggcta
|
gamma-glutamylcyclotransferase a |
| chr9_+_44430974 | 0.38 |
ENSDART00000056846
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr24_+_25471196 | 0.37 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr13_-_50139916 | 0.37 |
ENSDART00000099475
|
nid1a
|
nidogen 1a |
| chr1_+_8601935 | 0.36 |
ENSDART00000152367
|
si:ch211-160d14.6
|
si:ch211-160d14.6 |
| chr19_-_10394931 | 0.36 |
ENSDART00000191549
|
zgc:194578
|
zgc:194578 |
| chr6_-_20875111 | 0.36 |
ENSDART00000115118
ENSDART00000159916 |
tns1a
|
tensin 1a |
| chr21_-_22737228 | 0.35 |
ENSDART00000151366
|
fbxo40.2
|
F-box protein 40, tandem duplicate 2 |
| chr6_+_12462079 | 0.35 |
ENSDART00000192029
ENSDART00000065385 |
nr4a2b
|
nuclear receptor subfamily 4, group A, member 2b |
| chr23_+_17220986 | 0.35 |
ENSDART00000054761
|
nol4lb
|
nucleolar protein 4-like b |
| chr2_-_37797577 | 0.35 |
ENSDART00000110781
|
nfatc4
|
nuclear factor of activated T cells 4 |
| chr20_+_19066858 | 0.35 |
ENSDART00000192086
|
sox7
|
SRY (sex determining region Y)-box 7 |
| chr25_+_31222069 | 0.35 |
ENSDART00000159373
|
tnni2a.1
|
troponin I type 2a (skeletal, fast), tandem duplicate 1 |
| chr16_+_21330634 | 0.35 |
ENSDART00000191285
ENSDART00000183267 |
osbpl3b
|
oxysterol binding protein-like 3b |
| chr7_-_71758613 | 0.34 |
ENSDART00000166724
|
myom1b
|
myomesin 1b |
| chr10_-_24362775 | 0.34 |
ENSDART00000182104
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr23_-_40766518 | 0.34 |
ENSDART00000127420
|
si:dkeyp-27c8.2
|
si:dkeyp-27c8.2 |
| chr14_-_24332786 | 0.33 |
ENSDART00000173164
|
fam13b
|
family with sequence similarity 13, member B |
| chr12_-_26430507 | 0.33 |
ENSDART00000153214
|
synpo2lb
|
synaptopodin 2-like b |
| chr23_-_9925568 | 0.33 |
ENSDART00000081268
|
si:ch211-220i18.4
|
si:ch211-220i18.4 |
| chr21_-_25685739 | 0.33 |
ENSDART00000129619
ENSDART00000101205 |
phkg1b
|
phosphorylase kinase, gamma 1b (muscle) |
| chr6_+_8079974 | 0.32 |
ENSDART00000152071
|
si:ch211-207j7.2
|
si:ch211-207j7.2 |
| chr2_+_6885852 | 0.32 |
ENSDART00000016607
|
rgs5b
|
regulator of G protein signaling 5b |
| chr14_-_29906209 | 0.31 |
ENSDART00000192952
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr3_+_59784632 | 0.31 |
ENSDART00000084729
|
pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr3_+_18555899 | 0.31 |
ENSDART00000163036
ENSDART00000158791 |
cbx4
|
chromobox homolog 4 (Pc class homolog, Drosophila) |
| chr13_-_24379271 | 0.31 |
ENSDART00000046360
|
rhoua
|
ras homolog family member Ua |
| chr2_-_16254782 | 0.31 |
ENSDART00000133708
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr25_+_14092871 | 0.30 |
ENSDART00000067239
|
guca1g
|
guanylate cyclase activator 1g |
| chr3_+_53511936 | 0.30 |
ENSDART00000177083
|
zmp:0000001048
|
zmp:0000001048 |
| chr4_+_5848229 | 0.30 |
ENSDART00000161101
ENSDART00000067357 |
lyrm5a
|
LYR motif containing 5a |
| chr22_-_906757 | 0.30 |
ENSDART00000193182
|
FP016205.1
|
|
| chr2_+_6181383 | 0.30 |
ENSDART00000153307
|
si:ch73-344o19.1
|
si:ch73-344o19.1 |
| chr8_-_23758312 | 0.29 |
ENSDART00000132659
|
INAVA
|
si:ch211-163l21.4 |
| chr1_+_16396870 | 0.29 |
ENSDART00000189245
ENSDART00000113266 |
micu3a
|
mitochondrial calcium uptake family, member 3a |
| chr1_-_38813679 | 0.28 |
ENSDART00000148917
|
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr9_+_44430705 | 0.28 |
ENSDART00000190696
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr22_-_4769140 | 0.28 |
ENSDART00000165235
|
calr3a
|
calreticulin 3a |
| chr25_+_22107643 | 0.28 |
ENSDART00000089680
|
sigirr
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr25_+_10793478 | 0.27 |
ENSDART00000058339
ENSDART00000134923 |
ap3s2
|
adaptor-related protein complex 3, sigma 2 subunit |
| chr8_-_25002182 | 0.27 |
ENSDART00000078792
|
ahcyl1
|
adenosylhomocysteinase-like 1 |
| chr14_-_36799280 | 0.27 |
ENSDART00000168615
|
rnf130
|
ring finger protein 130 |
| chr14_+_31751260 | 0.27 |
ENSDART00000169796
|
si:dkeyp-11e3.1
|
si:dkeyp-11e3.1 |
| chr7_-_71758307 | 0.27 |
ENSDART00000161067
ENSDART00000165253 |
myom1b
|
myomesin 1b |
| chr12_-_13886952 | 0.27 |
ENSDART00000110503
|
adam11
|
ADAM metallopeptidase domain 11 |
| chr6_-_30859656 | 0.27 |
ENSDART00000156235
|
pde4ba
|
phosphodiesterase 4B, cAMP-specific a |
| chr4_+_11375894 | 0.26 |
ENSDART00000190471
ENSDART00000143963 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr14_-_32405387 | 0.26 |
ENSDART00000184647
|
fgf13a
|
fibroblast growth factor 13a |
| chr2_+_27403300 | 0.26 |
ENSDART00000099180
|
elovl8a
|
ELOVL fatty acid elongase 8a |
| chr11_+_6819050 | 0.26 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr11_+_25634041 | 0.26 |
ENSDART00000033657
|
grm6b
|
glutamate receptor, metabotropic 6b |
| chr24_+_34085940 | 0.25 |
ENSDART00000171189
|
asb10
|
ankyrin repeat and SOCS box containing 10 |
| chr3_-_30609659 | 0.25 |
ENSDART00000182516
ENSDART00000187047 ENSDART00000110597 |
syt3
|
synaptotagmin III |
| chr24_-_12745222 | 0.25 |
ENSDART00000151836
|
si:ch211-196f5.9
|
si:ch211-196f5.9 |
| chr2_+_16536453 | 0.25 |
ENSDART00000100287
|
grk7a
|
G protein-coupled receptor kinase 7a |
| chr23_-_21515182 | 0.24 |
ENSDART00000142000
|
rnf207b
|
ring finger protein 207b |
| chr12_+_10952794 | 0.24 |
ENSDART00000167090
|
raraa
|
retinoic acid receptor, alpha a |
| chr2_-_23768818 | 0.24 |
ENSDART00000148685
ENSDART00000191167 |
xirp1
|
xin actin binding repeat containing 1 |
| chr17_-_10249095 | 0.24 |
ENSDART00000159963
|
sstr1a
|
somatostatin receptor 1a |
| chr20_-_32332736 | 0.24 |
ENSDART00000062982
|
foxo3b
|
forkhead box O3b |
| chr17_-_43665366 | 0.24 |
ENSDART00000127945
|
egr2a
|
early growth response 2a |
| chr7_-_41964877 | 0.24 |
ENSDART00000092351
ENSDART00000193395 ENSDART00000187947 |
neto2b
|
neuropilin (NRP) and tolloid (TLL)-like 2b |
| chr18_-_8579907 | 0.23 |
ENSDART00000147284
|
FRMD4A
|
si:ch211-220f12.1 |
| chr12_+_34119439 | 0.22 |
ENSDART00000032821
|
cyth1b
|
cytohesin 1b |
| chr16_-_17200120 | 0.22 |
ENSDART00000147739
|
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr1_-_23040358 | 0.22 |
ENSDART00000146871
|
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr20_+_16881883 | 0.22 |
ENSDART00000130107
|
nfkbiaa
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a |
Network of associatons between targets according to the STRING database.
First level regulatory network of mef2ca+mef2cb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.2 | 0.5 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.2 | 2.8 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.2 | 0.6 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.2 | 0.9 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.2 | 0.9 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.2 | 0.7 | GO:1903019 | negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.2 | 0.5 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.2 | 0.9 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.2 | 0.5 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.2 | 0.8 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.2 | 1.5 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 0.8 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 0.4 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 1.9 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 1.3 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 | 1.4 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 0.6 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 0.7 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 0.2 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.1 | 0.6 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.5 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.7 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.1 | 1.2 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.1 | 0.3 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.1 | 0.4 | GO:0015709 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.1 | 0.8 | GO:0086003 | cardiac muscle cell contraction(GO:0086003) |
| 0.1 | 0.8 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.1 | 0.4 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) cellular response to vitamin D(GO:0071305) |
| 0.1 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 1.1 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.1 | 0.6 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.8 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.8 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 0.8 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 1.2 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.8 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 1.2 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.1 | GO:0046104 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.3 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.0 | 0.2 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.4 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 1.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.2 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.0 | 0.1 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.0 | 0.3 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.2 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.5 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.0 | 0.2 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 3.8 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.1 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.2 | GO:0043901 | negative regulation of multi-organism process(GO:0043901) |
| 0.0 | 0.6 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.8 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 1.8 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.2 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.3 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.2 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.0 | 0.8 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.5 | GO:0007622 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.0 | 0.4 | GO:1901642 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.9 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.3 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.4 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.1 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.0 | 0.5 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.9 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.6 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 1.6 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 0.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.3 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.3 | GO:0051560 | calcium ion transmembrane import into mitochondrion(GO:0036444) mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.5 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.5 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.6 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.8 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.2 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.1 | GO:1901571 | icosanoid transport(GO:0071715) fatty acid derivative transport(GO:1901571) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.3 | GO:0044773 | mitotic DNA damage checkpoint(GO:0044773) |
| 0.0 | 0.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.8 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 1.7 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.2 | 1.9 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.8 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 3.9 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.1 | 2.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.5 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 1.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 2.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.3 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.8 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.8 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 1.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.0 | GO:0032997 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.0 | 0.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.3 | 1.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.2 | 2.8 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.2 | 0.6 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.2 | 0.8 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.2 | 0.5 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.2 | 0.5 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.1 | 2.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.9 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 1.9 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 1.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.6 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.4 | GO:0004135 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.1 | 0.4 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 2.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.4 | GO:0015140 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 0.2 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.1 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.6 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.2 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 2.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.3 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.0 | 0.4 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.4 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.3 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.3 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.3 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.9 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.1 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.0 | 0.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 1.0 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.1 | GO:0002058 | nucleobase binding(GO:0002054) uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.0 | 1.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.3 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.1 | GO:0005372 | water transmembrane transporter activity(GO:0005372) |
| 0.0 | 0.9 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.2 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 5.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.3 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) structural constituent of synapse(GO:0098918) |
| 0.0 | 0.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 1.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.0 | GO:0019767 | immunoglobulin receptor activity(GO:0019763) IgE receptor activity(GO:0019767) |
| 0.0 | 0.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.4 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 0.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.2 | 0.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.5 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.8 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |