Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
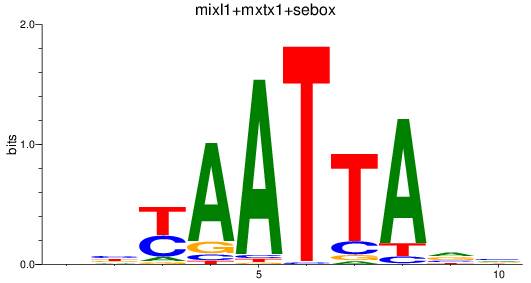
Results for mixl1+mxtx1+sebox
Z-value: 0.98
Transcription factors associated with mixl1+mxtx1+sebox
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sebox
|
ENSDARG00000042526 | SEBOX homeobox |
|
mixl1
|
ENSDARG00000069252 | Mix paired-like homeobox |
|
mxtx1
|
ENSDARG00000069382 | mix-type homeobox gene 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mixl1 | dr11_v1_chr20_-_43723860_43723860 | 0.49 | 1.8e-01 | Click! |
| mxtx1 | dr11_v1_chr13_-_21660203_21660203 | 0.42 | 2.6e-01 | Click! |
| sebox | dr11_v1_chr5_+_67371650_67371650 | -0.30 | 4.3e-01 | Click! |
Activity profile of mixl1+mxtx1+sebox motif
Sorted Z-values of mixl1+mxtx1+sebox motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_25777425 | 3.96 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr10_+_6884627 | 2.69 |
ENSDART00000125262
ENSDART00000121729 ENSDART00000105384 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr8_+_45334255 | 2.50 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr20_-_23426339 | 2.34 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr10_-_21362320 | 2.23 |
ENSDART00000189789
|
avd
|
avidin |
| chr10_-_21362071 | 2.21 |
ENSDART00000125167
|
avd
|
avidin |
| chr24_+_12835935 | 2.07 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr1_-_18811517 | 2.05 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr12_-_14143344 | 2.00 |
ENSDART00000152742
|
buc2l
|
bucky ball 2-like |
| chr16_-_29387215 | 1.99 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr18_-_40708537 | 1.89 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr2_+_6253246 | 1.82 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr1_-_55248496 | 1.74 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr14_+_34492288 | 1.68 |
ENSDART00000144301
|
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr10_-_25217347 | 1.56 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr14_+_34490445 | 1.53 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr10_+_6884123 | 1.47 |
ENSDART00000149095
ENSDART00000148772 ENSDART00000149334 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr24_+_1023839 | 1.46 |
ENSDART00000082526
|
zgc:111976
|
zgc:111976 |
| chr7_-_51773166 | 1.41 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr14_-_33481428 | 1.39 |
ENSDART00000147059
ENSDART00000140001 ENSDART00000124242 ENSDART00000164836 ENSDART00000190104 ENSDART00000186833 ENSDART00000180873 |
lamp2
|
lysosomal-associated membrane protein 2 |
| chr9_+_29548195 | 1.29 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr11_-_1550709 | 1.27 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr16_+_29509133 | 1.22 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr14_-_8940499 | 1.11 |
ENSDART00000129030
|
zgc:153681
|
zgc:153681 |
| chr1_+_35985813 | 1.06 |
ENSDART00000179634
ENSDART00000139636 ENSDART00000175902 |
zgc:152968
|
zgc:152968 |
| chr6_+_40922572 | 1.05 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr17_+_16046132 | 1.05 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr4_+_11723852 | 1.02 |
ENSDART00000028820
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr22_+_4488454 | 1.02 |
ENSDART00000170620
|
ctxn1
|
cortexin 1 |
| chr16_-_42056137 | 1.00 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr19_+_15441022 | 0.97 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr8_+_41037541 | 0.97 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr23_-_17003533 | 0.95 |
ENSDART00000080545
|
dnmt3bb.2
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.2 |
| chr2_+_41526904 | 0.94 |
ENSDART00000127520
|
acvr1l
|
activin A receptor, type 1 like |
| chr12_-_30359498 | 0.93 |
ENSDART00000152981
ENSDART00000189988 |
tdrd1
|
tudor domain containing 1 |
| chr3_+_18807006 | 0.91 |
ENSDART00000180091
|
tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr17_-_40956035 | 0.91 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr17_+_16046314 | 0.91 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr20_-_14114078 | 0.91 |
ENSDART00000168434
ENSDART00000104032 |
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr24_-_2450597 | 0.90 |
ENSDART00000188080
ENSDART00000093331 |
rreb1a
|
ras responsive element binding protein 1a |
| chr19_+_2631565 | 0.89 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr23_-_31913069 | 0.88 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr5_+_35786141 | 0.86 |
ENSDART00000022043
ENSDART00000127383 |
stard8
|
StAR-related lipid transfer (START) domain containing 8 |
| chr5_+_37903790 | 0.85 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr8_+_49117518 | 0.85 |
ENSDART00000079631
|
rad21l1
|
RAD21 cohesin complex component like 1 |
| chr7_+_34592526 | 0.84 |
ENSDART00000173959
|
fhod1
|
formin homology 2 domain containing 1 |
| chr20_-_37813863 | 0.84 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr16_-_17347727 | 0.79 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr24_-_30862168 | 0.79 |
ENSDART00000168540
|
ptbp2a
|
polypyrimidine tract binding protein 2a |
| chr11_+_35171406 | 0.78 |
ENSDART00000110839
|
mon1a
|
MON1 secretory trafficking family member A |
| chr5_-_20491198 | 0.76 |
ENSDART00000183051
ENSDART00000144232 |
ficd
|
FIC domain containing |
| chr21_-_36453594 | 0.75 |
ENSDART00000193176
|
cnot8
|
CCR4-NOT transcription complex, subunit 8 |
| chr16_+_47207691 | 0.75 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr19_+_15440841 | 0.74 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr6_-_40922971 | 0.72 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr5_+_6954162 | 0.69 |
ENSDART00000086666
|
stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr14_-_33945692 | 0.67 |
ENSDART00000168546
ENSDART00000189778 |
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr19_+_40069524 | 0.65 |
ENSDART00000151365
ENSDART00000140926 |
zmym4
|
zinc finger, MYM-type 4 |
| chr19_-_30510259 | 0.62 |
ENSDART00000135128
ENSDART00000186169 ENSDART00000182974 ENSDART00000187797 |
bag6l
|
BCL2 associated athanogene 6, like |
| chr3_+_26244353 | 0.62 |
ENSDART00000103733
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr10_-_33297864 | 0.61 |
ENSDART00000163360
|
PRDM15
|
PR/SET domain 15 |
| chr11_+_31864921 | 0.61 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr10_-_32494499 | 0.61 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr2_-_57076687 | 0.60 |
ENSDART00000161523
|
slc25a42
|
solute carrier family 25, member 42 |
| chr23_-_31913231 | 0.59 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr8_+_36500308 | 0.58 |
ENSDART00000098701
|
slc7a4
|
solute carrier family 7, member 4 |
| chr9_-_746317 | 0.58 |
ENSDART00000129632
ENSDART00000130720 |
usp37
|
ubiquitin specific peptidase 37 |
| chr3_-_30488063 | 0.56 |
ENSDART00000055393
ENSDART00000151367 |
med25
|
mediator complex subunit 25 |
| chr13_-_31017960 | 0.55 |
ENSDART00000145287
|
wdfy4
|
WDFY family member 4 |
| chr15_-_2519640 | 0.54 |
ENSDART00000047013
|
srprb
|
signal recognition particle receptor, B subunit |
| chr10_-_32494304 | 0.54 |
ENSDART00000028161
|
uvrag
|
UV radiation resistance associated gene |
| chr3_+_28860283 | 0.53 |
ENSDART00000077235
|
alg1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr13_+_1132261 | 0.53 |
ENSDART00000146049
|
wdr92
|
WD repeat domain 92 |
| chr1_-_29139141 | 0.53 |
ENSDART00000075546
ENSDART00000133246 |
hsf2bp
|
heat shock transcription factor 2 binding protein |
| chr1_-_45616470 | 0.53 |
ENSDART00000150165
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr18_+_19456648 | 0.53 |
ENSDART00000079695
|
zwilch
|
zwilch kinetochore protein |
| chr16_+_6750756 | 0.52 |
ENSDART00000149720
|
znf236
|
zinc finger protein 236 |
| chr21_-_25801956 | 0.52 |
ENSDART00000101219
|
mettl27
|
methyltransferase like 27 |
| chr23_-_25135046 | 0.51 |
ENSDART00000184844
ENSDART00000103989 |
idh3g
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr15_-_16177603 | 0.50 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr5_-_56412262 | 0.50 |
ENSDART00000083079
|
acaca
|
acetyl-CoA carboxylase alpha |
| chr8_+_11425048 | 0.50 |
ENSDART00000018739
|
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr24_-_24724233 | 0.50 |
ENSDART00000127044
ENSDART00000012399 |
armc1
|
armadillo repeat containing 1 |
| chr7_+_26649319 | 0.49 |
ENSDART00000173823
ENSDART00000101053 |
tp53i11a
|
tumor protein p53 inducible protein 11a |
| chr6_+_21001264 | 0.49 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr3_-_26244256 | 0.48 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr7_-_71389375 | 0.48 |
ENSDART00000128928
|
CABZ01074298.1
|
|
| chr2_+_20793982 | 0.47 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr11_-_35171162 | 0.46 |
ENSDART00000017393
|
traip
|
TRAF-interacting protein |
| chr1_+_21937201 | 0.45 |
ENSDART00000087729
|
kdm4c
|
lysine (K)-specific demethylase 4C |
| chr21_-_14174786 | 0.44 |
ENSDART00000145366
|
whrna
|
whirlin a |
| chr8_-_37249813 | 0.44 |
ENSDART00000098634
ENSDART00000140233 ENSDART00000061328 |
rbm39b
|
RNA binding motif protein 39b |
| chr5_+_4436405 | 0.43 |
ENSDART00000167969
|
CABZ01079241.1
|
|
| chr15_-_43284021 | 0.43 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr18_-_43884044 | 0.42 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr15_+_1534644 | 0.42 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr21_-_32060993 | 0.42 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr2_-_55298075 | 0.41 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr24_+_17068724 | 0.40 |
ENSDART00000191137
|
pip4k2aa
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha a |
| chr6_+_41191482 | 0.40 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr6_-_12172424 | 0.40 |
ENSDART00000109344
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr17_+_8799451 | 0.40 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr12_+_16087077 | 0.39 |
ENSDART00000141898
|
znf281b
|
zinc finger protein 281b |
| chr2_-_57378748 | 0.38 |
ENSDART00000149235
|
tcf3a
|
transcription factor 3a |
| chr20_-_45060241 | 0.38 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr17_-_41798856 | 0.37 |
ENSDART00000156031
ENSDART00000192801 ENSDART00000180172 ENSDART00000084745 ENSDART00000175577 |
ralgapa2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
| chr11_-_34522249 | 0.37 |
ENSDART00000158616
|
pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr16_+_25126935 | 0.37 |
ENSDART00000058945
|
zgc:92590
|
zgc:92590 |
| chr16_+_28994709 | 0.37 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr25_-_27621268 | 0.35 |
ENSDART00000146205
ENSDART00000073511 |
hyal6
|
hyaluronoglucosaminidase 6 |
| chr17_-_31639845 | 0.35 |
ENSDART00000154196
|
znf839
|
zinc finger protein 839 |
| chr15_-_4969525 | 0.33 |
ENSDART00000157223
|
lipt2
|
lipoyl(octanoyl) transferase 2 |
| chr14_+_44794936 | 0.33 |
ENSDART00000128881
|
zgc:195212
|
zgc:195212 |
| chr15_-_18115540 | 0.33 |
ENSDART00000131639
ENSDART00000047902 |
arcn1b
|
archain 1b |
| chr2_+_33326522 | 0.32 |
ENSDART00000056655
|
klf17
|
Kruppel-like factor 17 |
| chr15_+_31344472 | 0.32 |
ENSDART00000146695
ENSDART00000159182 ENSDART00000060125 |
or107-1
|
odorant receptor, family D, subfamily 107, member 1 |
| chr19_-_19379084 | 0.32 |
ENSDART00000165206
|
smarcc1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1b |
| chr8_+_50953776 | 0.31 |
ENSDART00000013870
|
zgc:56596
|
zgc:56596 |
| chr16_+_42471455 | 0.31 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr24_+_19415124 | 0.31 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr17_-_20118145 | 0.30 |
ENSDART00000149737
ENSDART00000165606 |
ryr2b
|
ryanodine receptor 2b (cardiac) |
| chr19_+_30884706 | 0.29 |
ENSDART00000052126
|
yars
|
tyrosyl-tRNA synthetase |
| chr21_-_11654422 | 0.29 |
ENSDART00000081614
ENSDART00000132699 |
cast
|
calpastatin |
| chr25_-_2723682 | 0.29 |
ENSDART00000113382
|
adpgk
|
ADP-dependent glucokinase |
| chr1_-_26444075 | 0.29 |
ENSDART00000125690
|
ints12
|
integrator complex subunit 12 |
| chr7_+_48667081 | 0.29 |
ENSDART00000083473
|
trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr2_-_3403020 | 0.29 |
ENSDART00000092741
|
snap47
|
synaptosomal-associated protein, 47 |
| chr8_+_2757821 | 0.28 |
ENSDART00000051403
ENSDART00000160551 |
sh3glb2a
|
SH3-domain GRB2-like endophilin B2a |
| chr7_+_23292133 | 0.28 |
ENSDART00000134489
|
htr2cl1
|
5-hydroxytryptamine (serotonin) receptor 2C, G protein-coupled-like 1 |
| chr11_+_34523132 | 0.27 |
ENSDART00000192257
|
zmat3
|
zinc finger, matrin-type 3 |
| chr14_+_8940326 | 0.27 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr6_+_3280939 | 0.27 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr17_-_16422654 | 0.27 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr2_-_32384683 | 0.26 |
ENSDART00000182942
ENSDART00000141757 |
ubtfl
|
upstream binding transcription factor, like |
| chr8_-_25033681 | 0.26 |
ENSDART00000003493
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr1_+_18811679 | 0.25 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr22_-_14739491 | 0.25 |
ENSDART00000133385
|
lrp1ba
|
low density lipoprotein receptor-related protein 1Ba |
| chr11_+_12052791 | 0.24 |
ENSDART00000158479
|
si:ch211-156l18.8
|
si:ch211-156l18.8 |
| chr5_+_36611128 | 0.24 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr5_-_37871526 | 0.24 |
ENSDART00000136450
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr8_-_37249991 | 0.24 |
ENSDART00000189275
ENSDART00000178556 |
rbm39b
|
RNA binding motif protein 39b |
| chr6_-_3982783 | 0.23 |
ENSDART00000171944
|
slc25a12
|
solute carrier family 25 (aspartate/glutamate carrier), member 12 |
| chr11_+_33818179 | 0.23 |
ENSDART00000109418
|
spoplb
|
speckle-type POZ protein-like b |
| chr3_-_16719244 | 0.23 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr8_-_16406440 | 0.22 |
ENSDART00000034004
|
faf1
|
Fas (TNFRSF6) associated factor 1 |
| chr24_+_7336411 | 0.21 |
ENSDART00000191170
|
kmt2ca
|
lysine (K)-specific methyltransferase 2Ca |
| chr13_+_22295905 | 0.21 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr21_-_25722834 | 0.20 |
ENSDART00000101208
|
abhd11
|
abhydrolase domain containing 11 |
| chr22_+_31207799 | 0.20 |
ENSDART00000133267
|
grip2b
|
glutamate receptor interacting protein 2b |
| chr1_+_8694196 | 0.20 |
ENSDART00000025604
|
zgc:77849
|
zgc:77849 |
| chr25_+_11456696 | 0.20 |
ENSDART00000171408
|
AGBL1
|
si:ch73-141f14.1 |
| chr3_-_31079186 | 0.19 |
ENSDART00000145636
ENSDART00000140569 |
ELOB (1 of many)
elob
|
elongin B elongin B |
| chr14_-_22113600 | 0.17 |
ENSDART00000113752
|
si:dkey-6i22.5
|
si:dkey-6i22.5 |
| chr20_-_45062514 | 0.17 |
ENSDART00000183529
ENSDART00000182955 |
klhl29
|
kelch-like family member 29 |
| chr20_-_7583486 | 0.16 |
ENSDART00000144729
|
usp24
|
ubiquitin specific peptidase 24 |
| chr17_+_37227936 | 0.15 |
ENSDART00000076009
|
hadhab
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha b |
| chr24_+_40860320 | 0.15 |
ENSDART00000161351
|
gorasp1b
|
golgi reassembly stacking protein 1b |
| chr11_+_18873113 | 0.14 |
ENSDART00000103969
ENSDART00000103968 |
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr10_+_40660772 | 0.14 |
ENSDART00000148007
|
taar19l
|
trace amine associated receptor 19l |
| chr16_-_16761164 | 0.14 |
ENSDART00000135872
|
si:dkey-27n14.1
|
si:dkey-27n14.1 |
| chr21_+_5272059 | 0.13 |
ENSDART00000141981
|
loxhd1a
|
lipoxygenase homology domains 1a |
| chr15_+_857148 | 0.12 |
ENSDART00000156949
|
si:dkey-7i4.13
|
si:dkey-7i4.13 |
| chr19_-_5103141 | 0.11 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr4_+_2655358 | 0.10 |
ENSDART00000007638
|
bcap29
|
B cell receptor associated protein 29 |
| chr21_+_34088377 | 0.10 |
ENSDART00000170070
|
mtmr1b
|
myotubularin related protein 1b |
| chr21_+_34088110 | 0.10 |
ENSDART00000145123
ENSDART00000029599 ENSDART00000147519 |
mtmr1b
|
myotubularin related protein 1b |
| chr4_-_5455506 | 0.10 |
ENSDART00000156593
ENSDART00000154676 |
si:dkey-14d8.22
|
si:dkey-14d8.22 |
| chr13_-_12602920 | 0.10 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr3_+_45364849 | 0.09 |
ENSDART00000153974
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr4_+_9177997 | 0.09 |
ENSDART00000057254
ENSDART00000154614 |
nfyba
|
nuclear transcription factor Y, beta a |
| chr3_+_45365098 | 0.09 |
ENSDART00000052746
ENSDART00000156555 |
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr8_+_25034544 | 0.08 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr23_+_39695827 | 0.07 |
ENSDART00000113893
ENSDART00000186679 |
tmco4
|
transmembrane and coiled-coil domains 4 |
| chr21_+_43404945 | 0.07 |
ENSDART00000142234
|
frmd7
|
FERM domain containing 7 |
| chr24_-_3783497 | 0.06 |
ENSDART00000158354
|
adarb2
|
adenosine deaminase, RNA-specific, B2 (non-functional) |
| chr3_+_32365811 | 0.06 |
ENSDART00000155967
|
ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr13_+_2625150 | 0.05 |
ENSDART00000164177
|
plpp4
|
phospholipid phosphatase 4 |
| chr15_+_34988148 | 0.05 |
ENSDART00000076269
|
ccdc105
|
coiled-coil domain containing 105 |
| chr8_+_23355484 | 0.05 |
ENSDART00000085361
ENSDART00000125729 |
dnmt3ba
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate a |
| chr20_-_5052786 | 0.05 |
ENSDART00000138818
ENSDART00000181655 ENSDART00000164274 |
arid1b
|
AT rich interactive domain 1B (SWI1-like) |
| chr23_+_31912882 | 0.05 |
ENSDART00000140505
|
armc1l
|
armadillo repeat containing 1, like |
| chr21_+_15592426 | 0.05 |
ENSDART00000138207
|
smarcb1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1b |
| chr23_+_33963619 | 0.04 |
ENSDART00000140666
ENSDART00000084792 |
plpbp
|
pyridoxal phosphate binding protein |
| chr24_+_32668675 | 0.04 |
ENSDART00000156638
ENSDART00000155973 |
si:ch211-282b22.1
|
si:ch211-282b22.1 |
| chr8_+_37527575 | 0.04 |
ENSDART00000147239
|
or135-1
|
odorant receptor, family H, subfamily 135, member 1 |
| chr11_+_29770966 | 0.03 |
ENSDART00000088624
ENSDART00000124471 |
rpgrb
|
retinitis pigmentosa GTPase regulator b |
| chr13_+_25486608 | 0.02 |
ENSDART00000057689
|
bag3
|
BCL2 associated athanogene 3 |
| chr12_+_48803098 | 0.01 |
ENSDART00000074768
|
ppifb
|
peptidylprolyl isomerase Fb |
| chr16_+_33902006 | 0.00 |
ENSDART00000161807
ENSDART00000159474 |
gnl2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
Network of associatons between targets according to the STRING database.
First level regulatory network of mixl1+mxtx1+sebox
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0039015 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.8 | 4.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.5 | 1.5 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.4 | 1.7 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.4 | 1.4 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.3 | 4.2 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.2 | 1.0 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.2 | 0.9 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.2 | 0.9 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.2 | 2.8 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 1.4 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.9 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.1 | 1.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.5 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.1 | 0.3 | GO:2000374 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.1 | 0.3 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.1 | 0.4 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 0.9 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 0.9 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 0.3 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.1 | 1.1 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.7 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 0.6 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.5 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.5 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.1 | 0.2 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.3 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.1 | 0.3 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 2.6 | GO:0060968 | regulation of gene silencing(GO:0060968) |
| 0.0 | 0.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.2 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.0 | 0.3 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 1.5 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.3 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 3.4 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.8 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.7 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 0.4 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.1 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.4 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.3 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.6 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.4 | GO:0048696 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.2 | GO:0015810 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.4 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.8 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.0 | 0.5 | GO:0072599 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.0 | 0.5 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.5 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 2.0 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 0.4 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.7 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.6 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.2 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.9 | GO:0044744 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.2 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 | 0.3 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.9 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.5 | GO:0007006 | mitochondrial membrane organization(GO:0007006) |
| 0.0 | 0.4 | GO:0048634 | regulation of muscle organ development(GO:0048634) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.5 | 1.4 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.2 | 6.8 | GO:0043186 | P granule(GO:0043186) |
| 0.2 | 0.9 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.1 | 0.9 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 0.9 | GO:0034991 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 2.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.7 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.4 | GO:0002141 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.0 | 0.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.2 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 2.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.4 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 4.6 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.7 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.8 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0035060 | brahma complex(GO:0035060) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.4 | GO:0009374 | biotin binding(GO:0009374) |
| 0.3 | 1.5 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 1.4 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.2 | 0.6 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.2 | 2.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 2.8 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 2.5 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 0.5 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 1.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.9 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.3 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
| 0.1 | 0.5 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.4 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 0.3 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.1 | 0.9 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 1.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 2.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.3 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 4.6 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 0.7 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.2 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 1.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 0.6 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.8 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.4 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 1.7 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.1 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.0 | 0.4 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 6.0 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 1.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 1.1 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 4.2 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 3.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 2.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.6 | PID CDC42 PATHWAY | CDC42 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.5 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |