Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
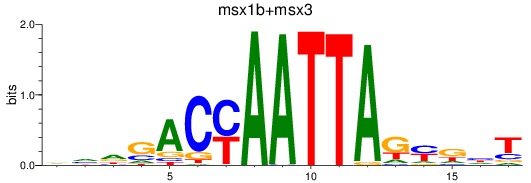
Results for msx1b+msx3
Z-value: 0.27
Transcription factors associated with msx1b+msx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
msx1b
|
ENSDARG00000008886 | muscle segment homeobox 1b |
|
msx3
|
ENSDARG00000015674 | muscle segment homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| msx1b | dr11_v1_chr1_+_49352900_49352900 | 0.75 | 2.0e-02 | Click! |
| msx3 | dr11_v1_chr13_+_24662238_24662238 | 0.43 | 2.5e-01 | Click! |
Activity profile of msx1b+msx3 motif
Sorted Z-values of msx1b+msx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_19402802 | 0.34 |
ENSDART00000135552
|
rbm47
|
RNA binding motif protein 47 |
| chr14_+_9421510 | 0.29 |
ENSDART00000123652
|
hmgn6
|
high mobility group nucleosome binding domain 6 |
| chr24_+_22485710 | 0.29 |
ENSDART00000146058
|
si:dkey-40h20.1
|
si:dkey-40h20.1 |
| chr24_+_20373605 | 0.24 |
ENSDART00000112349
|
plcd1a
|
phospholipase C, delta 1a |
| chr8_-_15129573 | 0.24 |
ENSDART00000142358
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr21_-_20328375 | 0.24 |
ENSDART00000079593
|
slc26a1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr8_+_26293673 | 0.22 |
ENSDART00000144977
|
mgll
|
monoglyceride lipase |
| chr15_-_20024205 | 0.22 |
ENSDART00000161379
|
auts2b
|
autism susceptibility candidate 2b |
| chr13_+_30903816 | 0.21 |
ENSDART00000191727
|
ercc6
|
excision repair cross-complementation group 6 |
| chr1_-_58036509 | 0.20 |
ENSDART00000081122
|
COLGALT1
|
si:ch211-114l13.7 |
| chr17_+_24821627 | 0.20 |
ENSDART00000112389
|
wdr43
|
WD repeat domain 43 |
| chr6_+_50451337 | 0.18 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr20_+_25568694 | 0.17 |
ENSDART00000063107
ENSDART00000063128 |
cyp2p7
|
cytochrome P450, family 2, subfamily P, polypeptide 7 |
| chr15_+_46344655 | 0.16 |
ENSDART00000155893
|
si:ch1073-340i21.2
|
si:ch1073-340i21.2 |
| chr21_-_37973081 | 0.15 |
ENSDART00000136569
|
ripply1
|
ripply transcriptional repressor 1 |
| chr20_+_11731039 | 0.14 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr14_+_34490445 | 0.13 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr15_-_14212777 | 0.13 |
ENSDART00000165572
|
CR925813.1
|
|
| chr15_-_25518084 | 0.13 |
ENSDART00000158594
|
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr21_+_15592426 | 0.13 |
ENSDART00000138207
|
smarcb1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1b |
| chr6_+_39370587 | 0.12 |
ENSDART00000157165
ENSDART00000155079 |
si:dkey-195m11.8
|
si:dkey-195m11.8 |
| chr4_+_54519511 | 0.12 |
ENSDART00000161653
|
znf974
|
zinc finger protein 974 |
| chr25_+_19666621 | 0.12 |
ENSDART00000112746
|
zgc:171459
|
zgc:171459 |
| chr4_-_20292821 | 0.12 |
ENSDART00000136069
ENSDART00000192504 |
cacna2d4a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4a |
| chr11_-_11910225 | 0.11 |
ENSDART00000159922
|
si:ch211-69b7.6
|
si:ch211-69b7.6 |
| chr11_+_24716837 | 0.11 |
ENSDART00000145217
|
zgc:153953
|
zgc:153953 |
| chr3_+_34140507 | 0.11 |
ENSDART00000131802
|
si:dkey-204f11.64
|
si:dkey-204f11.64 |
| chr3_+_23692462 | 0.10 |
ENSDART00000145934
|
hoxb7a
|
homeobox B7a |
| chr23_-_16485190 | 0.10 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr9_+_21277846 | 0.10 |
ENSDART00000139620
ENSDART00000110996 ENSDART00000111899 |
lats2
|
large tumor suppressor kinase 2 |
| chr3_-_31079186 | 0.10 |
ENSDART00000145636
ENSDART00000140569 |
ELOB (1 of many)
elob
|
elongin B elongin B |
| chr5_-_29534748 | 0.10 |
ENSDART00000159587
|
AL831768.1
|
|
| chr17_+_24851951 | 0.10 |
ENSDART00000180746
|
cx35.4
|
connexin 35.4 |
| chr5_+_27897504 | 0.10 |
ENSDART00000130936
|
adam28
|
ADAM metallopeptidase domain 28 |
| chr21_+_723998 | 0.09 |
ENSDART00000160956
|
oaz1b
|
ornithine decarboxylase antizyme 1b |
| chr11_+_37251825 | 0.09 |
ENSDART00000169804
|
il17rc
|
interleukin 17 receptor C |
| chr13_+_18321140 | 0.09 |
ENSDART00000180947
|
eif4e1c
|
eukaryotic translation initiation factor 4E family member 1c |
| chr2_+_16487443 | 0.09 |
ENSDART00000114980
|
CR377211.1
|
|
| chr2_+_10771787 | 0.08 |
ENSDART00000187782
|
gfi1aa
|
growth factor independent 1A transcription repressor a |
| chr18_+_9171778 | 0.07 |
ENSDART00000101192
|
sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr10_+_36178713 | 0.07 |
ENSDART00000140816
|
or108-3
|
odorant receptor, family D, subfamily 108, member 3 |
| chr15_+_5360407 | 0.07 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr6_-_9695294 | 0.07 |
ENSDART00000162728
|
nop58
|
NOP58 ribonucleoprotein homolog (yeast) |
| chr12_+_3078221 | 0.07 |
ENSDART00000148835
ENSDART00000149427 |
sgca
|
sarcoglycan, alpha |
| chr2_+_6253246 | 0.07 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr16_+_35595312 | 0.07 |
ENSDART00000170438
|
si:ch211-1i11.3
|
si:ch211-1i11.3 |
| chr14_-_1543834 | 0.06 |
ENSDART00000185876
|
CABZ01109480.1
|
|
| chr1_+_55730205 | 0.06 |
ENSDART00000148075
|
adgre10
|
adhesion G protein-coupled receptor E10 |
| chr9_+_35832294 | 0.06 |
ENSDART00000130549
ENSDART00000122169 |
si:dkey-67c22.2
|
si:dkey-67c22.2 |
| chr4_-_67969695 | 0.06 |
ENSDART00000190016
|
si:ch211-223k15.1
|
si:ch211-223k15.1 |
| chr1_+_54626491 | 0.06 |
ENSDART00000136063
|
si:ch211-202h22.9
|
si:ch211-202h22.9 |
| chr4_+_77047067 | 0.06 |
ENSDART00000174206
|
znf1009
|
zinc finger protein 1009 |
| chr10_+_39212898 | 0.06 |
ENSDART00000159501
|
stt3a
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr15_-_37719015 | 0.05 |
ENSDART00000154875
|
si:dkey-117a8.1
|
si:dkey-117a8.1 |
| chr10_-_44560165 | 0.05 |
ENSDART00000181217
ENSDART00000076084 |
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr18_+_50461981 | 0.05 |
ENSDART00000158761
|
CU896640.1
|
|
| chr4_-_12997348 | 0.05 |
ENSDART00000171451
|
si:dkey-6a5.3
|
si:dkey-6a5.3 |
| chr10_+_39212601 | 0.04 |
ENSDART00000168778
|
stt3a
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr22_+_2769236 | 0.04 |
ENSDART00000141836
|
si:dkey-20i20.10
|
si:dkey-20i20.10 |
| chr4_-_42408339 | 0.04 |
ENSDART00000172612
|
si:ch211-59d8.3
|
si:ch211-59d8.3 |
| chr4_-_62714083 | 0.04 |
ENSDART00000188299
|
si:dkey-28k24.2
|
si:dkey-28k24.2 |
| chr16_-_7379328 | 0.04 |
ENSDART00000060447
|
med18
|
mediator of RNA polymerase II transcription, subunit 18 homolog (yeast) |
| chr7_+_8324506 | 0.04 |
ENSDART00000168110
|
si:dkey-185m8.2
|
si:dkey-185m8.2 |
| chr15_-_19051152 | 0.04 |
ENSDART00000186453
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr23_+_44427188 | 0.04 |
ENSDART00000160207
|
eif4e2rs1
|
eukaryotic translation initiation factor 4E family member 2 related sequence 1 |
| chr12_-_31457301 | 0.03 |
ENSDART00000043887
ENSDART00000148603 |
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr2_-_20788698 | 0.03 |
ENSDART00000181823
|
CABZ01020455.1
|
|
| chr5_+_872299 | 0.03 |
ENSDART00000130042
|
fubp3
|
far upstream element (FUSE) binding protein 3 |
| chr4_-_9891874 | 0.03 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr7_-_8374950 | 0.03 |
ENSDART00000057101
|
aep1
|
aerolysin-like protein |
| chr17_+_26352372 | 0.03 |
ENSDART00000155177
|
grid1a
|
glutamate receptor, ionotropic, delta 1a |
| chr24_+_26345609 | 0.03 |
ENSDART00000186844
|
lrrc34
|
leucine rich repeat containing 34 |
| chr22_+_9761930 | 0.03 |
ENSDART00000181107
|
BX664625.5
|
|
| chr10_+_36194527 | 0.03 |
ENSDART00000076810
|
or108-1
|
odorant receptor, family D, subfamily 108, member 1 |
| chr25_+_16116740 | 0.03 |
ENSDART00000139778
|
far1
|
fatty acyl CoA reductase 1 |
| chr24_+_26345427 | 0.03 |
ENSDART00000089756
|
lrrc34
|
leucine rich repeat containing 34 |
| chr10_+_21867307 | 0.03 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr5_-_47727819 | 0.03 |
ENSDART00000165249
|
cox7c
|
cytochrome c oxidase, subunit VIIc |
| chr16_-_26855936 | 0.03 |
ENSDART00000167320
ENSDART00000078119 |
ino80c
|
INO80 complex subunit C |
| chr21_-_22648007 | 0.03 |
ENSDART00000121788
|
gig2l
|
grass carp reovirus (GCRV)-induced gene 2l |
| chr7_-_8324927 | 0.03 |
ENSDART00000102535
|
f13a1b
|
coagulation factor XIII, A1 polypeptide b |
| chr17_+_7513673 | 0.02 |
ENSDART00000156674
|
klhl10b.1
|
kelch-like family member 10b, tandem duplicate 1 |
| chr4_+_62576422 | 0.02 |
ENSDART00000180679
|
si:ch211-79g12.2
|
si:ch211-79g12.2 |
| chr11_-_6974022 | 0.02 |
ENSDART00000172851
|
COMP
|
si:ch211-43f4.1 |
| chr15_+_36188171 | 0.02 |
ENSDART00000156598
|
masp1
|
mannan-binding lectin serine peptidase 1 |
| chr10_+_40568735 | 0.02 |
ENSDART00000136468
ENSDART00000182841 |
taar18i
|
trace amine associated receptor 18i |
| chr16_+_33144112 | 0.02 |
ENSDART00000183149
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr15_-_37683768 | 0.02 |
ENSDART00000059609
|
si:dkey-117a8.3
|
si:dkey-117a8.3 |
| chr24_+_17345521 | 0.02 |
ENSDART00000024722
ENSDART00000154250 |
ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr22_-_26274177 | 0.02 |
ENSDART00000060978
|
wdr83
|
WD repeat domain containing 83 |
| chr21_-_8085635 | 0.02 |
ENSDART00000082790
|
si:dkey-163m14.2
|
si:dkey-163m14.2 |
| chr15_-_37709072 | 0.02 |
ENSDART00000157115
|
si:dkey-117a8.2
|
si:dkey-117a8.2 |
| chr11_-_25538341 | 0.02 |
ENSDART00000171560
|
si:dkey-245f22.3
|
si:dkey-245f22.3 |
| chr7_-_69647988 | 0.02 |
ENSDART00000169943
|
LO018231.1
|
|
| chr25_+_10947743 | 0.02 |
ENSDART00000073381
|
mhc1lfa
|
major histocompatibility complex class I LFA |
| chr5_+_7564644 | 0.02 |
ENSDART00000192173
|
CABZ01039096.1
|
|
| chr10_+_11355841 | 0.02 |
ENSDART00000193067
ENSDART00000064215 |
cops4
|
COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis) |
| chr24_-_25004553 | 0.02 |
ENSDART00000080997
ENSDART00000136860 |
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr6_-_19042294 | 0.02 |
ENSDART00000159461
|
si:rp71-81e14.2
|
si:rp71-81e14.2 |
| chr10_+_17235370 | 0.02 |
ENSDART00000038780
|
sppl3
|
signal peptide peptidase 3 |
| chr16_+_33144306 | 0.02 |
ENSDART00000101953
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr15_-_37620167 | 0.01 |
ENSDART00000059582
|
si:ch211-137j23.8
|
si:ch211-137j23.8 |
| chr21_-_38730557 | 0.01 |
ENSDART00000150984
ENSDART00000111885 |
taf9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr3_+_2719243 | 0.01 |
ENSDART00000189256
|
CR388047.3
|
|
| chr5_+_4806851 | 0.01 |
ENSDART00000067599
|
angptl2a
|
angiopoietin-like 2a |
| chr15_-_37673733 | 0.01 |
ENSDART00000154513
|
si:dkey-117a8.4
|
si:dkey-117a8.4 |
| chr12_-_18883079 | 0.01 |
ENSDART00000178619
|
shisa8b
|
shisa family member 8b |
| chr22_+_18816662 | 0.01 |
ENSDART00000132476
|
cbarpb
|
calcium channel, voltage-dependent, beta subunit associated regulatory protein b |
| chr2_+_30369116 | 0.01 |
ENSDART00000142137
|
pi15b
|
peptidase inhibitor 15b |
| chr13_+_27328098 | 0.01 |
ENSDART00000037585
|
mb21d1
|
Mab-21 domain containing 1 |
| chr8_-_53680653 | 0.01 |
ENSDART00000164739
|
wnt5a
|
wingless-type MMTV integration site family, member 5a |
| chr4_-_75681004 | 0.01 |
ENSDART00000186296
|
si:dkey-71l4.5
|
si:dkey-71l4.5 |
| chr16_+_5774977 | 0.01 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr10_-_40416054 | 0.01 |
ENSDART00000137947
ENSDART00000143606 |
taar20j
|
trace amine associated receptor 20j |
| chr1_+_40801448 | 0.01 |
ENSDART00000187394
|
cpz
|
carboxypeptidase Z |
| chr9_-_35633827 | 0.01 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr14_+_6535426 | 0.01 |
ENSDART00000055961
|
thg1l
|
tRNA-histidine guanylyltransferase 1-like |
| chr17_+_15041647 | 0.01 |
ENSDART00000108999
|
samd4a
|
sterile alpha motif domain containing 4A |
| chr22_+_9114416 | 0.01 |
ENSDART00000190169
|
nlrp15
|
NACHT, LRR and PYD domains-containing protein 15 |
| chr15_+_31303355 | 0.01 |
ENSDART00000143423
|
or116-2
|
odorant receptor, family F, subfamily 116, member 2 |
| chr22_+_5532003 | 0.01 |
ENSDART00000106174
|
si:ch73-256j6.2
|
si:ch73-256j6.2 |
| chr15_-_37733238 | 0.01 |
ENSDART00000115178
|
si:dkey-42l23.7
|
si:dkey-42l23.7 |
| chr6_+_52877147 | 0.01 |
ENSDART00000174370
|
or137-8
|
odorant receptor, family H, subfamily 137, member 8 |
| chr8_-_27656765 | 0.01 |
ENSDART00000078491
|
mov10b.2
|
Moloney leukemia virus 10b, tandem duplicate 2 |
| chr22_+_1615691 | 0.01 |
ENSDART00000171555
|
si:ch211-255f4.13
|
si:ch211-255f4.13 |
| chr11_+_42726712 | 0.00 |
ENSDART00000028955
|
tdrd3
|
tudor domain containing 3 |
| chr25_+_6471401 | 0.00 |
ENSDART00000132927
|
snx33
|
sorting nexin 33 |
| chr18_+_34225520 | 0.00 |
ENSDART00000126115
|
v2rl1
|
vomeronasal 2 receptor, l1 |
| chr3_+_32663865 | 0.00 |
ENSDART00000190240
|
si:dkey-16l2.16
|
si:dkey-16l2.16 |
| chr7_+_24496894 | 0.00 |
ENSDART00000149994
|
nelfa
|
negative elongation factor complex member A |
| chr24_-_9979342 | 0.00 |
ENSDART00000138576
ENSDART00000191206 |
zgc:171977
|
zgc:171977 |
| chr20_-_47188966 | 0.00 |
ENSDART00000152965
|
si:dkeyp-104f11.9
|
si:dkeyp-104f11.9 |
| chr6_-_18971063 | 0.00 |
ENSDART00000188163
|
sept9b
|
septin 9b |
| chr3_-_31715714 | 0.00 |
ENSDART00000051542
|
ccdc47
|
coiled-coil domain containing 47 |
| chr8_-_7567815 | 0.00 |
ENSDART00000132536
|
hcfc1b
|
host cell factor C1b |
| chr15_-_37767901 | 0.00 |
ENSDART00000154763
|
si:dkey-42l23.4
|
si:dkey-42l23.4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of msx1b+msx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:2000742 | regulation of anterior head development(GO:2000742) |
| 0.1 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.2 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.1 | GO:0007508 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 0.0 | 0.0 | GO:0010747 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.0 | 0.2 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0035060 | brahma complex(GO:0035060) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.0 | GO:0048030 | disaccharide binding(GO:0048030) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |