Project
PRJNA195909:zebrafish embryo and larva development
Navigation
Downloads
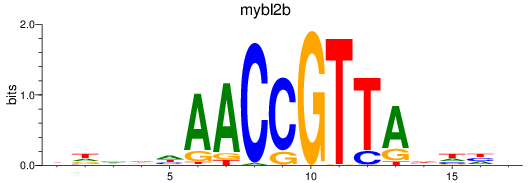
Results for mybl2b
Z-value: 1.93
Transcription factors associated with mybl2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mybl2b
|
ENSDARG00000032264 | v-myb avian myeloblastosis viral oncogene homolog-like 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mybl2b | dr11_v1_chr11_+_1565806_1565806 | 0.88 | 1.8e-03 | Click! |
Activity profile of mybl2b motif
Sorted Z-values of mybl2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_43656125 | 4.95 |
ENSDART00000136392
ENSDART00000192805 ENSDART00000181960 |
si:ch211-263m18.4
si:dkey-229d11.5
|
si:ch211-263m18.4 si:dkey-229d11.5 |
| chr10_-_26202766 | 4.78 |
ENSDART00000136393
|
fhdc3
|
FH2 domain containing 3 |
| chr2_+_6253246 | 4.46 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr2_+_307665 | 3.52 |
ENSDART00000082083
|
zgc:113452
|
zgc:113452 |
| chr19_+_43579786 | 2.72 |
ENSDART00000138404
|
si:ch211-199g17.2
|
si:ch211-199g17.2 |
| chr8_-_1730686 | 2.54 |
ENSDART00000183802
|
CABZ01065416.1
|
|
| chr25_-_16552135 | 2.44 |
ENSDART00000125836
|
si:ch211-266k8.4
|
si:ch211-266k8.4 |
| chr1_-_354115 | 2.40 |
ENSDART00000141590
ENSDART00000098627 |
pros1
|
protein S |
| chr24_-_21150100 | 2.37 |
ENSDART00000154249
ENSDART00000193256 |
gramd1c
|
GRAM domain containing 1c |
| chr16_-_41714988 | 2.20 |
ENSDART00000138798
|
cep85
|
centrosomal protein 85 |
| chr10_-_35257458 | 2.18 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr18_-_399554 | 2.14 |
ENSDART00000164374
ENSDART00000186311 ENSDART00000181816 ENSDART00000181892 |
si:ch211-79l17.1
|
si:ch211-79l17.1 |
| chr19_-_46566430 | 2.14 |
ENSDART00000166668
|
ext1b
|
exostosin glycosyltransferase 1b |
| chr10_-_45029041 | 2.14 |
ENSDART00000167878
|
polm
|
polymerase (DNA directed), mu |
| chr9_+_18829360 | 2.07 |
ENSDART00000006514
|
gtf2f2b
|
general transcription factor IIF, polypeptide 2b |
| chr10_+_16501699 | 2.06 |
ENSDART00000121864
|
slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr14_+_1365097 | 2.02 |
ENSDART00000083797
ENSDART00000176087 |
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr9_+_22782027 | 1.95 |
ENSDART00000090816
|
rif1
|
replication timing regulatory factor 1 |
| chr16_+_52343905 | 1.90 |
ENSDART00000131051
|
ifnlr1
|
interferon lambda receptor 1 |
| chr2_-_51507540 | 1.88 |
ENSDART00000166605
ENSDART00000161093 |
pigrl2.3
|
polymeric immunoglobulin receptor-like 2.3 |
| chr13_+_45524475 | 1.85 |
ENSDART00000074567
ENSDART00000019113 |
maco1b
|
macoilin 1b |
| chr9_-_2892045 | 1.81 |
ENSDART00000137201
|
cdca7a
|
cell division cycle associated 7a |
| chr8_+_26226044 | 1.78 |
ENSDART00000186503
|
wdr6
|
WD repeat domain 6 |
| chr6_+_2195625 | 1.78 |
ENSDART00000155659
|
acvr1bb
|
activin A receptor type 1Bb |
| chr17_-_40879108 | 1.75 |
ENSDART00000010362
|
supt7l
|
SPT7-like STAGA complex gamma subunit |
| chr16_+_25066880 | 1.72 |
ENSDART00000154084
|
im:7147486
|
im:7147486 |
| chr16_+_42667560 | 1.71 |
ENSDART00000023452
|
dpy19l1l
|
dpy-19-like 1, like (H. sapiens) |
| chr3_+_1211242 | 1.66 |
ENSDART00000171287
ENSDART00000165769 |
poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr4_-_837768 | 1.63 |
ENSDART00000185280
ENSDART00000135618 |
sobpb
|
sine oculis binding protein homolog (Drosophila) b |
| chr4_-_72643171 | 1.61 |
ENSDART00000130126
|
CABZ01054394.1
|
|
| chr9_-_29003245 | 1.58 |
ENSDART00000183391
ENSDART00000188836 |
ptpn4a
|
protein tyrosine phosphatase, non-receptor type 4a |
| chr4_-_1684335 | 1.58 |
ENSDART00000019144
ENSDART00000113360 |
arid2
|
AT rich interactive domain 2 (ARID, RFX-like) |
| chr10_+_43406796 | 1.58 |
ENSDART00000184337
ENSDART00000183034 ENSDART00000180623 ENSDART00000132486 |
rasa1b
|
RAS p21 protein activator (GTPase activating protein) 1b |
| chr11_+_25583950 | 1.54 |
ENSDART00000111961
|
ccdc120
|
coiled-coil domain containing 120 |
| chr20_+_13781779 | 1.54 |
ENSDART00000142999
ENSDART00000152471 |
lpgat1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr16_-_31342356 | 1.53 |
ENSDART00000136648
|
mroh1
|
maestro heat-like repeat family member 1 |
| chr8_+_23382568 | 1.53 |
ENSDART00000129167
|
mapre1a
|
microtubule-associated protein, RP/EB family, member 1a |
| chr5_+_59234421 | 1.51 |
ENSDART00000045518
|
prkrip1
|
PRKR interacting protein 1 |
| chr2_+_38025260 | 1.49 |
ENSDART00000075905
|
hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr3_+_13929860 | 1.49 |
ENSDART00000164179
|
syce2
|
synaptonemal complex central element protein 2 |
| chr9_-_2892250 | 1.44 |
ENSDART00000140695
|
cdca7a
|
cell division cycle associated 7a |
| chr24_-_25166416 | 1.43 |
ENSDART00000111552
ENSDART00000169495 |
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr25_+_35134393 | 1.41 |
ENSDART00000185379
|
CR762436.2
|
|
| chr5_+_25084385 | 1.38 |
ENSDART00000134526
ENSDART00000111863 |
paxx
|
PAXX, non-homologous end joining factor |
| chr17_-_45378473 | 1.31 |
ENSDART00000132969
|
znf106a
|
zinc finger protein 106a |
| chr7_+_5909271 | 1.31 |
ENSDART00000173363
|
zgc:165555
|
zgc:165555 |
| chr10_+_35257651 | 1.31 |
ENSDART00000028940
|
styxl1
|
serine/threonine/tyrosine interacting-like 1 |
| chr21_+_6212844 | 1.29 |
ENSDART00000150301
|
fnbp1b
|
formin binding protein 1b |
| chr6_-_9563534 | 1.28 |
ENSDART00000184765
|
map3k2
|
mitogen-activated protein kinase kinase kinase 2 |
| chr7_+_1579236 | 1.27 |
ENSDART00000172830
|
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr21_-_1635268 | 1.26 |
ENSDART00000151258
|
zgc:152948
|
zgc:152948 |
| chr24_-_41180149 | 1.24 |
ENSDART00000019975
|
acvr2ba
|
activin A receptor type 2Ba |
| chr10_-_16868211 | 1.23 |
ENSDART00000171755
|
stoml2
|
stomatin (EPB72)-like 2 |
| chr3_-_9444749 | 1.22 |
ENSDART00000182191
|
FO904885.3
|
|
| chr24_-_25166720 | 1.22 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr18_+_3579829 | 1.22 |
ENSDART00000158763
ENSDART00000182850 ENSDART00000162754 ENSDART00000178789 ENSDART00000172656 |
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr10_+_3145707 | 1.22 |
ENSDART00000160046
|
hic2
|
hypermethylated in cancer 2 |
| chr4_-_2380173 | 1.21 |
ENSDART00000177727
|
nap1l1
|
nucleosome assembly protein 1-like 1 |
| chr3_+_32080403 | 1.19 |
ENSDART00000124943
|
aldh16a1
|
aldehyde dehydrogenase 16 family, member A1 |
| chr17_-_31819837 | 1.19 |
ENSDART00000160281
|
abraxas2b
|
abraxas 2b, BRISC complex subunit |
| chr12_+_4220353 | 1.19 |
ENSDART00000133675
|
mapk7
|
mitogen-activated protein kinase 7 |
| chr24_-_12832499 | 1.17 |
ENSDART00000182027
ENSDART00000039312 |
ipo4
|
importin 4 |
| chr22_+_10166407 | 1.17 |
ENSDART00000092104
|
pxk
|
PX domain containing serine/threonine kinase |
| chr4_-_5831036 | 1.17 |
ENSDART00000166232
|
foxm1
|
forkhead box M1 |
| chr20_+_42563618 | 1.15 |
ENSDART00000153441
|
igf2r
|
insulin-like growth factor 2 receptor |
| chr4_-_26035770 | 1.14 |
ENSDART00000124514
|
usp44
|
ubiquitin specific peptidase 44 |
| chr2_-_2451181 | 1.14 |
ENSDART00000101033
|
pth1ra
|
parathyroid hormone 1 receptor a |
| chr18_-_18587745 | 1.10 |
ENSDART00000191973
|
sf3b3
|
splicing factor 3b, subunit 3 |
| chr19_+_4968947 | 1.09 |
ENSDART00000003634
ENSDART00000134808 |
stard3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr8_-_12432604 | 1.08 |
ENSDART00000133350
ENSDART00000140699 ENSDART00000101174 |
traf1
|
TNF receptor-associated factor 1 |
| chr2_-_37632896 | 1.08 |
ENSDART00000008302
|
insra
|
insulin receptor a |
| chr9_+_28140089 | 1.04 |
ENSDART00000046880
|
plekhm3
|
pleckstrin homology domain containing, family M, member 3 |
| chr18_-_41160579 | 1.04 |
ENSDART00000181480
|
CABZ01005876.1
|
|
| chr4_+_72205923 | 1.03 |
ENSDART00000181075
|
zgc:165515
|
zgc:165515 |
| chr3_-_33941319 | 1.00 |
ENSDART00000026090
ENSDART00000111878 |
gtf2f1
|
general transcription factor IIF, polypeptide 1 |
| chr5_+_6796291 | 0.99 |
ENSDART00000166868
ENSDART00000165308 |
me2
|
malic enzyme 2, NAD(+)-dependent, mitochondrial |
| chr13_+_15701849 | 0.97 |
ENSDART00000003517
|
trmt61a
|
tRNA methyltransferase 61A |
| chr2_-_193707 | 0.97 |
ENSDART00000187642
|
znf1014
|
zinc finger protein 1014 |
| chr19_+_16068445 | 0.96 |
ENSDART00000138709
|
gmeb1
|
glucocorticoid modulatory element binding protein 1 |
| chr17_-_43012390 | 0.95 |
ENSDART00000155615
|
atg2b
|
autophagy related 2B |
| chr2_-_1622641 | 0.95 |
ENSDART00000082143
|
prkacbb
|
protein kinase, cAMP-dependent, catalytic, beta b |
| chr2_-_22286828 | 0.95 |
ENSDART00000168653
|
fam110b
|
family with sequence similarity 110, member B |
| chr7_-_17337233 | 0.94 |
ENSDART00000050236
ENSDART00000102141 |
nitr8
|
novel immune-type receptor 8 |
| chr7_-_6352569 | 0.93 |
ENSDART00000173074
|
zgc:165555
|
zgc:165555 |
| chr25_+_3099073 | 0.93 |
ENSDART00000022506
|
rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr6_+_3473657 | 0.90 |
ENSDART00000011785
|
rad54l
|
RAD54 like |
| chr3_+_14641962 | 0.90 |
ENSDART00000091070
|
zgc:158403
|
zgc:158403 |
| chr12_-_7280551 | 0.90 |
ENSDART00000061633
|
zgc:171971
|
zgc:171971 |
| chr21_+_39432248 | 0.90 |
ENSDART00000179938
|
pafah1b1b
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1b |
| chr20_+_21595244 | 0.89 |
ENSDART00000010643
|
esr2a
|
estrogen receptor 2a |
| chr10_+_42542517 | 0.89 |
ENSDART00000005496
|
kctd9b
|
potassium channel tetramerization domain containing 9b |
| chr17_-_11466700 | 0.88 |
ENSDART00000091159
|
adpgk2
|
ADP-dependent glucokinase 2 |
| chr17_-_22573311 | 0.87 |
ENSDART00000141523
ENSDART00000140022 ENSDART00000079390 ENSDART00000188644 |
exo1
|
exonuclease 1 |
| chr22_+_24157807 | 0.87 |
ENSDART00000159165
|
b3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr3_-_33941875 | 0.87 |
ENSDART00000047660
|
gtf2f1
|
general transcription factor IIF, polypeptide 1 |
| chr8_+_23381892 | 0.86 |
ENSDART00000180950
ENSDART00000063010 ENSDART00000074241 ENSDART00000142783 |
mapre1a
|
microtubule-associated protein, RP/EB family, member 1a |
| chr1_+_23559478 | 0.85 |
ENSDART00000133659
|
ncapg
|
non-SMC condensin I complex, subunit G |
| chr25_-_13403726 | 0.84 |
ENSDART00000056723
|
gins3
|
GINS complex subunit 3 |
| chr11_-_20988238 | 0.84 |
ENSDART00000155238
|
taf4a
|
TAF4A RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr5_-_61588998 | 0.83 |
ENSDART00000050912
|
pex12
|
peroxisomal biogenesis factor 12 |
| chr21_+_34119759 | 0.82 |
ENSDART00000024750
ENSDART00000128242 |
hmgb3b
|
high mobility group box 3b |
| chr2_+_10006839 | 0.82 |
ENSDART00000160304
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr25_+_34938317 | 0.81 |
ENSDART00000042678
|
vps4a
|
vacuolar protein sorting 4a homolog A (S. cerevisiae) |
| chr22_+_1431743 | 0.80 |
ENSDART00000182871
|
BX322657.2
|
Danio rerio si:dkeyp-53d3.6 (si:dkeyp-53d3.6), mRNA. |
| chr6_-_36795111 | 0.79 |
ENSDART00000160669
ENSDART00000104256 ENSDART00000187751 ENSDART00000161928 ENSDART00000183264 |
opa1
|
optic atrophy 1 (autosomal dominant) |
| chr25_-_37319117 | 0.79 |
ENSDART00000044499
|
igl4v9
|
immunoglobulin light 4 variable 9 |
| chr19_-_47571456 | 0.77 |
ENSDART00000158071
ENSDART00000165841 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr2_+_10007113 | 0.77 |
ENSDART00000155213
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr25_-_3192405 | 0.76 |
ENSDART00000104835
|
hps5
|
Hermansky-Pudlak syndrome 5 |
| chr1_-_23293261 | 0.76 |
ENSDART00000122648
|
ugdh
|
UDP-glucose 6-dehydrogenase |
| chr2_-_9696283 | 0.75 |
ENSDART00000165712
|
selenot1a
|
selenoprotein T, 1a |
| chr23_+_36308717 | 0.75 |
ENSDART00000131711
|
hnrnpa1b
|
heterogeneous nuclear ribonucleoprotein A1b |
| chr11_-_40519886 | 0.75 |
ENSDART00000172819
|
miip
|
migration and invasion inhibitory protein |
| chr5_-_1869982 | 0.73 |
ENSDART00000055878
|
rcl1
|
RNA terminal phosphate cyclase-like 1 |
| chr20_+_53181017 | 0.71 |
ENSDART00000189692
ENSDART00000177109 |
FIG4
|
FIG4 phosphoinositide 5-phosphatase |
| chr20_+_28266892 | 0.70 |
ENSDART00000103330
|
chac1
|
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr11_+_5588122 | 0.67 |
ENSDART00000113281
|
zgc:172302
|
zgc:172302 |
| chr7_+_17106160 | 0.65 |
ENSDART00000190048
ENSDART00000180004 ENSDART00000013409 |
prmt3
|
protein arginine methyltransferase 3 |
| chr15_-_31508221 | 0.64 |
ENSDART00000121464
|
si:dkey-1m11.5
|
si:dkey-1m11.5 |
| chr8_-_16675464 | 0.64 |
ENSDART00000191982
|
osbpl9
|
oxysterol binding protein-like 9 |
| chr20_-_26047741 | 0.62 |
ENSDART00000141626
ENSDART00000112714 ENSDART00000152102 ENSDART00000078535 ENSDART00000151874 |
gtf2h5
|
general transcription factor IIH, polypeptide 5 |
| chr20_-_52271262 | 0.62 |
ENSDART00000135463
|
rhpn1
|
rhophilin, Rho GTPase binding protein 1 |
| chr10_+_43037064 | 0.62 |
ENSDART00000160159
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr7_-_33130552 | 0.61 |
ENSDART00000127006
|
mns1
|
meiosis-specific nuclear structural 1 |
| chr8_-_25761544 | 0.61 |
ENSDART00000078152
|
suv39h1b
|
suppressor of variegation 3-9 homolog 1b |
| chr20_-_22798794 | 0.61 |
ENSDART00000148084
|
fip1l1a
|
FIP1 like 1a (S. cerevisiae) |
| chr11_-_24015845 | 0.59 |
ENSDART00000191783
|
chia.3
|
chitinase, acidic.3 |
| chr12_+_34953038 | 0.58 |
ENSDART00000187022
ENSDART00000123988 ENSDART00000027034 |
qki2
|
QKI, KH domain containing, RNA binding 2 |
| chr6_-_26080384 | 0.53 |
ENSDART00000157181
ENSDART00000154568 |
hs2st1b
|
heparan sulfate 2-O-sulfotransferase 1b |
| chr15_+_17074185 | 0.53 |
ENSDART00000046648
|
clptm1
|
cleft lip and palate associated transmembrane protein 1 |
| chr7_-_24994722 | 0.53 |
ENSDART00000131671
|
rcor2
|
REST corepressor 2 |
| chr22_-_8006342 | 0.52 |
ENSDART00000162028
|
sc:d217
|
sc:d217 |
| chr16_-_24708586 | 0.52 |
ENSDART00000178614
|
BX005256.1
|
|
| chr6_+_29693663 | 0.52 |
ENSDART00000150083
|
pde6d
|
phosphodiesterase 6D, cGMP-specific, rod, delta |
| chr15_-_38218320 | 0.51 |
ENSDART00000152363
|
rhoga
|
ras homolog family member Ga |
| chr7_+_19738665 | 0.49 |
ENSDART00000089333
|
tkfc
|
triokinase/FMN cyclase |
| chr8_-_47206114 | 0.49 |
ENSDART00000147606
ENSDART00000060883 ENSDART00000060880 |
ppih
|
peptidylprolyl isomerase H (cyclophilin H) |
| chr8_+_47571211 | 0.49 |
ENSDART00000131460
|
plch2a
|
phospholipase C, eta 2a |
| chr22_+_21153031 | 0.48 |
ENSDART00000166342
|
crlf1b
|
cytokine receptor-like factor 1b |
| chr14_-_4043818 | 0.48 |
ENSDART00000179870
|
snx25
|
sorting nexin 25 |
| chr25_-_5815834 | 0.47 |
ENSDART00000112853
|
appl2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr12_-_27242498 | 0.46 |
ENSDART00000152609
ENSDART00000152170 |
si:dkey-11c5.11
|
si:dkey-11c5.11 |
| chr18_+_910992 | 0.46 |
ENSDART00000161206
ENSDART00000167229 |
pkma
|
pyruvate kinase M1/2a |
| chr9_+_14023942 | 0.46 |
ENSDART00000142749
|
si:ch211-67e16.4
|
si:ch211-67e16.4 |
| chr22_+_28803739 | 0.45 |
ENSDART00000129476
ENSDART00000189726 |
tp53bp2b
|
tumor protein p53 binding protein, 2b |
| chr21_-_22681534 | 0.45 |
ENSDART00000159233
|
gig2f
|
grass carp reovirus (GCRV)-induced gene 2f |
| chr1_+_30946231 | 0.45 |
ENSDART00000022841
|
metap1d
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr20_-_52271015 | 0.45 |
ENSDART00000074307
|
rhpn1
|
rhophilin, Rho GTPase binding protein 1 |
| chr21_+_37090795 | 0.45 |
ENSDART00000085786
|
znf346
|
zinc finger protein 346 |
| chr22_+_21152847 | 0.44 |
ENSDART00000007321
|
crlf1b
|
cytokine receptor-like factor 1b |
| chr18_-_50676567 | 0.44 |
ENSDART00000172264
|
ube2q1
|
ubiquitin-conjugating enzyme E2Q family member 1 |
| chr8_+_16676894 | 0.44 |
ENSDART00000076586
|
si:ch211-198n5.11
|
si:ch211-198n5.11 |
| chr1_-_8020589 | 0.41 |
ENSDART00000143881
|
si:dkeyp-9d4.2
|
si:dkeyp-9d4.2 |
| chr16_-_7214513 | 0.41 |
ENSDART00000150005
|
si:ch73-341k19.1
|
si:ch73-341k19.1 |
| chr5_-_32882162 | 0.40 |
ENSDART00000085769
|
lrsam1
|
leucine rich repeat and sterile alpha motif containing 1 |
| chr23_-_10722664 | 0.39 |
ENSDART00000146526
ENSDART00000129022 ENSDART00000104985 |
foxp1a
|
forkhead box P1a |
| chr6_+_29693492 | 0.38 |
ENSDART00000114172
|
pde6d
|
phosphodiesterase 6D, cGMP-specific, rod, delta |
| chr5_+_42951852 | 0.38 |
ENSDART00000097615
|
mob1ba
|
MOB kinase activator 1Ba |
| chr5_-_61797220 | 0.37 |
ENSDART00000079855
|
im:7138535
|
im:7138535 |
| chr8_-_47206399 | 0.37 |
ENSDART00000060877
|
ppih
|
peptidylprolyl isomerase H (cyclophilin H) |
| chr9_-_36924388 | 0.36 |
ENSDART00000059756
|
ralba
|
v-ral simian leukemia viral oncogene homolog Ba (ras related) |
| chr15_-_17074393 | 0.35 |
ENSDART00000155526
|
si:ch211-24o10.6
|
si:ch211-24o10.6 |
| chr22_+_2801464 | 0.33 |
ENSDART00000132419
|
si:dkey-20i20.7
|
si:dkey-20i20.7 |
| chr22_+_14117078 | 0.33 |
ENSDART00000013575
|
bzw1a
|
basic leucine zipper and W2 domains 1a |
| chr8_+_25629941 | 0.33 |
ENSDART00000186925
|
wasa
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia) a |
| chr12_+_19138452 | 0.33 |
ENSDART00000141346
ENSDART00000066397 |
phf5a
|
PHD finger protein 5A |
| chr9_+_14023386 | 0.32 |
ENSDART00000140199
ENSDART00000124267 |
si:ch211-67e16.4
|
si:ch211-67e16.4 |
| chr18_+_1145571 | 0.32 |
ENSDART00000135055
|
rec114
|
REC114 meiotic recombination protein |
| chr24_+_5912635 | 0.32 |
ENSDART00000153736
|
pimr63
|
Pim proto-oncogene, serine/threonine kinase, related 63 |
| chr2_+_25315591 | 0.32 |
ENSDART00000161386
|
fndc3ba
|
fibronectin type III domain containing 3Ba |
| chr20_-_51267185 | 0.31 |
ENSDART00000158350
ENSDART00000023064 |
slc35b2
|
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B2 |
| chr18_+_29898955 | 0.30 |
ENSDART00000064080
|
cenpn
|
centromere protein N |
| chr22_+_2306386 | 0.30 |
ENSDART00000148300
ENSDART00000147644 |
si:dkey-4c15.16
|
si:dkey-4c15.16 |
| chr17_+_1632294 | 0.30 |
ENSDART00000191959
|
srp14
|
signal recognition particle 14 |
| chr10_+_6121558 | 0.30 |
ENSDART00000166799
ENSDART00000157947 |
tln1
|
talin 1 |
| chr3_+_23248542 | 0.30 |
ENSDART00000185765
ENSDART00000192332 |
ppp1r9ba
|
protein phosphatase 1, regulatory subunit 9Ba |
| chr7_-_24994568 | 0.29 |
ENSDART00000002961
|
rcor2
|
REST corepressor 2 |
| chr4_-_48578324 | 0.29 |
ENSDART00000122707
|
znf980
|
zinc finger protein 980 |
| chr2_+_9970942 | 0.29 |
ENSDART00000153063
|
sec22ba
|
SEC22 homolog B, vesicle trafficking protein a |
| chr24_-_42108655 | 0.29 |
ENSDART00000032722
|
pigm
|
phosphatidylinositol glycan anchor biosynthesis, class M |
| chr24_+_24809877 | 0.27 |
ENSDART00000185117
|
dnajc5b
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr9_+_37320222 | 0.27 |
ENSDART00000100622
|
slc15a2
|
solute carrier family 15 (oligopeptide transporter), member 2 |
| chr12_-_26491464 | 0.26 |
ENSDART00000153361
|
si:dkey-287g12.6
|
si:dkey-287g12.6 |
| chr6_-_33875919 | 0.24 |
ENSDART00000190411
|
tmem69
|
transmembrane protein 69 |
| chr10_-_26985231 | 0.22 |
ENSDART00000050585
|
znhit2
|
zinc finger, HIT-type containing 2 |
| chr22_-_23590069 | 0.19 |
ENSDART00000172067
|
f13b
|
coagulation factor XIII, B polypeptide |
| chr17_-_45383699 | 0.17 |
ENSDART00000141182
|
tmem206
|
transmembrane protein 206 |
| chr12_+_48803098 | 0.16 |
ENSDART00000074768
|
ppifb
|
peptidylprolyl isomerase Fb |
| chr18_-_18821929 | 0.16 |
ENSDART00000178875
|
aars
|
alanyl-tRNA synthetase |
| chr4_+_308697 | 0.15 |
ENSDART00000174021
ENSDART00000131419 ENSDART00000067489 |
si:ch211-51e12.7
|
si:ch211-51e12.7 |
| chr18_-_26797723 | 0.14 |
ENSDART00000008013
|
sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr20_-_46817223 | 0.14 |
ENSDART00000100336
|
esrrgb
|
estrogen-related receptor gamma b |
| chr4_-_54100377 | 0.11 |
ENSDART00000188064
|
si:dkey-199m13.7
|
si:dkey-199m13.7 |
| chr9_-_28275600 | 0.11 |
ENSDART00000170094
|
creb1b
|
cAMP responsive element binding protein 1b |
| chr19_+_31904836 | 0.10 |
ENSDART00000162297
ENSDART00000088340 ENSDART00000151280 ENSDART00000151218 |
tpd52
|
tumor protein D52 |
| chr17_-_35352690 | 0.09 |
ENSDART00000016053
|
rnf144aa
|
ring finger protein 144aa |
| chr24_-_36337395 | 0.08 |
ENSDART00000154402
|
si:ch211-40k21.9
|
si:ch211-40k21.9 |
| chr15_+_5277761 | 0.08 |
ENSDART00000153954
|
si:ch1073-166e24.4
|
si:ch1073-166e24.4 |
| chr20_-_35052464 | 0.08 |
ENSDART00000037195
|
kif26bb
|
kinesin family member 26Bb |
| chr23_+_20689255 | 0.07 |
ENSDART00000182420
|
usp21
|
ubiquitin specific peptidase 21 |
Network of associatons between targets according to the STRING database.
First level regulatory network of mybl2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.6 | 1.7 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.4 | 1.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.4 | 1.5 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.4 | 1.9 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.3 | 1.3 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.3 | 1.1 | GO:1904667 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.3 | 2.4 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.3 | 4.5 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 0.9 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 3.1 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.1 | 3.5 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.1 | 0.8 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 0.5 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.1 | 0.8 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.1 | 0.8 | GO:0003188 | heart valve formation(GO:0003188) atrioventricular valve formation(GO:0003190) |
| 0.1 | 1.0 | GO:1902534 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) single-organism membrane invagination(GO:1902534) |
| 0.1 | 1.6 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 1.0 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.7 | GO:1902315 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.1 | 2.7 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 0.9 | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.1 | 0.9 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 0.7 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 1.2 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.1 | 3.2 | GO:0048538 | thymus development(GO:0048538) |
| 0.1 | 1.5 | GO:0007130 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.1 | 0.5 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.1 | 2.7 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.1 | 3.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 0.8 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 0.6 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.7 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.1 | 0.9 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 | 0.9 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 2.2 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 1.1 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.1 | 1.6 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.1 | 0.4 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 0.9 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 3.0 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.1 | 0.7 | GO:0000479 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.1 | 1.1 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 | 0.6 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.1 | 1.8 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.9 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 2.1 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.0 | 0.8 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.7 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.8 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.5 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 2.2 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 1.5 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 1.3 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.7 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 4.5 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.9 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 1.9 | GO:0000723 | telomere maintenance(GO:0000723) telomere organization(GO:0032200) |
| 0.0 | 1.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 1.1 | GO:0030301 | cholesterol transport(GO:0030301) |
| 0.0 | 0.8 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 1.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.3 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 1.4 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 1.2 | GO:0007041 | lysosomal transport(GO:0007041) |
| 0.0 | 0.5 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.1 | GO:0032475 | otolith formation(GO:0032475) |
| 0.0 | 0.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.5 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 1.2 | GO:0044744 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 1.9 | GO:0048884 | neuromast development(GO:0048884) |
| 0.0 | 0.6 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 3.0 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.3 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 1.9 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.5 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.0 | 0.8 | GO:0016575 | histone deacetylation(GO:0016575) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.9 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.3 | 0.9 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.3 | 0.8 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.3 | 1.8 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.2 | 1.5 | GO:0000801 | central element(GO:0000801) |
| 0.2 | 0.9 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.2 | 0.9 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.2 | 1.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 3.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.2 | 1.0 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.2 | 1.4 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.2 | 0.8 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 1.1 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 1.1 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.1 | 2.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 1.3 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 0.9 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 2.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.6 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 1.9 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 0.3 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 0.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 1.9 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.8 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 1.0 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.8 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.0 | 1.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.5 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 2.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 2.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 2.7 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 1.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.5 | 1.9 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 0.4 | 4.5 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.3 | 1.8 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.3 | 2.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.3 | 0.9 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.3 | 0.8 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.3 | 0.8 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.2 | 1.0 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.2 | 1.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.2 | 1.1 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.2 | 0.6 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.2 | 1.0 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.2 | 0.9 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.2 | 0.8 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.2 | 0.5 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.2 | 0.7 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.2 | 1.6 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.2 | 0.9 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.9 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.1 | 0.4 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.1 | 0.7 | GO:0090554 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 1.2 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 1.0 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.1 | 0.8 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.3 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 3.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 2.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 1.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.8 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 0.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 1.2 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 0.6 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 2.0 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 1.0 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.9 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 1.5 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.1 | 0.6 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.8 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 6.7 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.3 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.7 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.9 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 1.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.7 | GO:0032934 | sterol binding(GO:0032934) |
| 0.0 | 1.2 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.9 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 2.8 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.8 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 1.6 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 4.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.7 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 2.2 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 2.1 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 2.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 1.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.1 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 1.7 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.8 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 1.5 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 1.9 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 1.3 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 2.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 1.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.1 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.8 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.8 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.8 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.3 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |